This is a preprint.
Genome-wide Association Identifies Novel Etiological Insights Associated with Parkinson's Disease in African and African Admixed Populations
- PMID: 37398408
- PMCID: PMC10312852
- DOI: 10.1101/2023.05.05.23289529
Genome-wide Association Identifies Novel Etiological Insights Associated with Parkinson's Disease in African and African Admixed Populations
Update in
-
Identification of genetic risk loci and causal insights associated with Parkinson's disease in African and African admixed populations: a genome-wide association study.Lancet Neurol. 2023 Nov;22(11):1015-1025. doi: 10.1016/S1474-4422(23)00283-1. Epub 2023 Aug 23. Lancet Neurol. 2023. PMID: 37633302 Free PMC article.
Abstract
Background: Understanding the genetic mechanisms underlying diseases in ancestrally diverse populations is a critical step towards the realization of the global application of precision medicine. The African and African admixed populations enable mapping of complex traits given their greater levels of genetic diversity, extensive population substructure, and distinct linkage disequilibrium patterns.
Methods: Here we perform a comprehensive genome-wide assessment of Parkinson's disease (PD) in 197,918 individuals (1,488 cases; 196,430 controls) of African and African admixed ancestry, characterizing population-specific risk, differential haplotype structure and admixture, coding and structural genetic variation and polygenic risk profiling.
Findings: We identified a novel common risk factor for PD and age at onset at the GBA1 locus (risk, rs3115534-G; OR=1.58, 95% CI = 1.37 - 1.80, P=2.397E-14; age at onset, BETA =-2.004, SE =0.57, P = 0.0005), that was found to be rare in non-African/African admixed populations. Downstream short- and long-read whole genome sequencing analyses did not reveal any coding or structural variant underlying the GWAS signal. However, we identified that this signal mediates PD risk via expression quantitative trait locus (eQTL) mechanisms. While previously identified GBA1 associated disease risk variants are coding mutations, here we suggest a novel functional mechanism consistent with a trend in decreasing glucocerebrosidase activity levels. Given the high population frequency of the underlying signal and the phenotypic characteristics of the homozygous carriers, we hypothesize that this variant may not cause Gaucher disease. Additionally, the prevalence of Gaucher's disease in Africa is low.
Interpretation: The present study identifies a novel African-ancestry genetic risk factor in GBA1 as a major mechanistic basis of PD in the African and African admixed populations. This striking result contrasts to previous work in Northern European populations, both in terms of mechanism and attributable risk. This finding highlights the importance of understanding population-specific genetic risk in complex diseases, a particularly crucial point as the field moves toward precision medicine in PD clinical trials and while recognizing the need for equitable inclusion of ancestrally diverse groups in such trials. Given the distinctive genetics of these underrepresented populations, their inclusion represents a valuable step towards insights into novel genetic determinants underlying PD etiology. This opens new avenues towards RNA-based and other therapeutic strategies aimed at reducing lifetime risk.
Evidence before this study: Our current understanding of Parkinson's disease (PD) is disproportionately based on studying populations of European ancestry, leading to a significant gap in our knowledge about the genetics, clinical characteristics, and pathophysiology in underrepresented populations. This is particularly notable in individuals of African and African admixed ancestries. Over the last two decades, we have witnessed a revolution in the research area of complex genetic diseases. In the PD field, large-scale genome-wide association studies in the European, Asian, and Latin American populations have identified multiple risk loci associated with disease. These include 78 loci and 90 independent signals associated with PD risk in the European population, nine replicated loci and two novel population-specific signals in the Asian population, and a total of 11 novel loci recently nominated through multi-ancestry GWAS efforts.Nevertheless, the African and African admixed populations remain completely unexplored in the context of PD genetics.
Added value of this study: To address the lack of diversity in our research field, this study aimed to conduct the first genome-wide assessment of PD genetics in the African and African admixed populations. Here, we identified a genetic risk factor linked to PD etiology, dissected African-specific differences in risk and age at onset, characterized known genetic risk factors, and highlighted the utility of the African and African admixed risk haplotype substructure for future fine-mapping efforts. We identified a novel disease mechanism via expression changes consistent with decreased GBA1 activity levels. Future large scale single cell expression studies should investigate the neuronal populations in which expression differences are most prominent. This novel mechanism may hold promise for future efficient RNA-based therapeutic strategies such as antisense oligonucleotides or short interfering RNAs aimed at preventing and decreasing disease risk. We envisage that these data generated under the umbrella of the Global Parkinson's Genetics Program (GP2) will shed light on the molecular mechanisms involved in the disease process and might pave the way for future clinical trials and therapeutic interventions. This work represents a valuable resource in an underserved population, supporting pioneering research within GP2 and beyond. Deciphering causal and genetic risk factors in all these ancestries will help determine whether interventions, potential targets for disease modifying treatment, and prevention strategies that are being studied in the European populations are relevant to the African and African admixed populations.
Implications of all the available evidence: We nominate a novel signal impacting GBA1 as the major genetic risk factor for PD in the African and African admixed populations. The present study could inform future GBA1 clinical trials, improving patient stratification. In this regard, genetic testing can help to design trials likely to provide meaningful and actionable answers. It is our hope that these findings may ultimately have clinical utility for this underrepresented population.
Conflict of interest statement
This research was supported in part by the Intramural Research Program of the NIH, National Institute on Aging (NIA), National Institutes of Health, Department of Health and Human Services; project number ZO1 AG000535 and ZIA AG000949, as well as the National Institute of Neurological Disorders and Stroke (NINDS) and the National Human Genome Research Institute (NHGRI).
This work was supported in part by the Global Parkinson’s Genetics Program (GP2). GP2 is funded by the Aligning Science Against Parkinson’s (ASAP) initiative and implemented by The Michael J. Fox Foundation for Parkinson’s Research (
D.V., H.I., H.L.L., K.L. and M.A.N.’s participation in this project was part of a competitive contract awarded to Data Tecnica International LLC by the National Institutes of Health to support open science research. K.H. and members of the 23andMe Research Team are employed by and hold stock or stock options in 23andMe, Inc. M.A.N. also currently serves on the scientific advisory board for Character Biosciences Inc and Neuron 23 Inc.
DGS is a member of the faculty of the University of Alabama at Birmingham and is supported by endowment and University funds, is an investigator in studies funded by Abbvie, Inc., the American Parkinson Disease Association, the Michael J. Fox Foundation for Parkinson Research, The National Parkinson Foundation, Alabama Department of Commerce, Alabama Innovation Fund, Genentech, the Department of Defense, and NIH grants P50NS108675 and R25NS079188 and has a clinical practice and is compensated for these activities through the University of Alabama Health Services Foundation. He serves as Deputy Editor for the journal Movement Disorders and is compensated for this role by the International Parkinson and Movement Disorders Society. In addition, since January 1, 2022 he has served as a consultant for or received honoraria from Abbvie Inc., Curium Pharma, Appello, Theravance, Sanofi-Aventis, Alnylam Pharmaceutics, Coave Therapeutics, BlueRock Therapeutics and F. Hoffman-La Roche.
We thank the research participants and employees of 23andMe. The following members of the 23andMe Research Team contributed to this study:
Stella Aslibekyan, Adam Auton, Elizabeth Babalola, Robert K. Bell, Jessica Bielenberg, Katarzyna Bryc, Emily Bullis, Paul Cannon, Daniella Coker, Gabriel Cuellar Partida, Devika Dhamija, Sayantan Das, Sarah L. Elson, Nicholas Eriksson, Teresa Filshtein, Alison Fitch, Kipper Fletez-Brant, Pierre Fontanillas, Will Freyman, Julie M. Granka, Alejandro Hernandez, Barry Hicks, David A. Hinds, Ethan M. Jewett, Yunxuan Jiang, Katelyn Kukar, Alan Kwong, Keng-Han Lin, Bianca A. Llamas, Maya Lowe, Jey C. McCreight, Matthew H. McIntyre, Steven J. Micheletti, Meghan E. Moreno, Priyanka Nandakumar, Dominique T. Nguyen, Elizabeth S. Noblin, Jared O’Connell, Aaron A. Petrakovitz, G. David Poznik, Alexandra Reynoso, Madeleine Schloetter, Morgan Schumacher, Anjali J. Shastri, Janie F. Shelton, Jingchunzi Shi, Suyash Shringarpure, Qiaojuan Jane Su, Susana A. Tat, Christophe Toukam Tchakouté, Vinh Tran, Joyce Y. Tung, Xin Wang, Wei Wang, Catherine H. Weldon, Peter Wilton, Corinna D. Wong
We thank Cynthia J. Casaceli, Debbie Baker and Christi Alessi-Fox from the University of Rochester Clinical Trials Coordination Center for its contribution to the coordination of the BLAAC PD Study. We thank Lisa Shulman for her contribution to the design of the protocol for BLAAC PD clinical assessments.
We thank the Biowulf team, as this study used the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health (
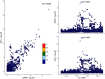
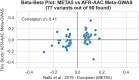
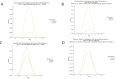
Figure 1 was generated on
Figures




References
-
- Chang M, He L, Cai L. An Overview of Genome-Wide Association Studies. Methods Mol Biol 2018; 1754: 97–108. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
