This is a preprint.
The patient-specific mouse model with Foxg1 frameshift mutation uncovers the pathophysiology of FOXG1 syndrome
- PMID: 37398410
- PMCID: PMC10312924
- DOI: 10.21203/rs.3.rs-2953760/v1
The patient-specific mouse model with Foxg1 frameshift mutation uncovers the pathophysiology of FOXG1 syndrome
Update in
-
The patient-specific mouse model with Foxg1 frameshift mutation provides insights into the pathophysiology of FOXG1 syndrome.Nat Commun. 2025 May 22;16(1):4760. doi: 10.1038/s41467-025-59838-4. Nat Commun. 2025. PMID: 40404610 Free PMC article.
Abstract
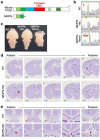
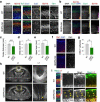
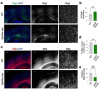
Single allelic mutations in the gene encoding the forebrain-specific transcription factor FOXG1 lead to FOXG1 syndrome (FS). Patient-specific animal models are needed to understand the etiology of FS, as FS patients show a wide spectrum of symptoms correlated with location and mutation type in the FOXG1 gene. Here we report the first patient-specific FS mouse model, Q84Pfs heterozygous (Q84Pfs-Het) mice, mimicking one of the most predominant single nucleotide variants in FS. Intriguingly, we found that Q84Pfs-Het mice faithfully recapitulate human FS phenotypes at the cellular, brain structural, and behavioral levels. Importantly, Q84Pfs-Het mice exhibited myelination deficits like FS patients. Further, our transcriptome analysis of Q84Pfs-Het cortex revealed a new role for FOXG1 in synapse and oligodendrocyte development. The dysregulated genes in Q84Pfs-Het brains also predicted motor dysfunction and autism-like phenotypes. Correspondingly, Q84Pfs-Het mice showed movement deficits, repetitive behaviors, increased anxiety, and prolonged behavior arrest. Together, our study revealed the crucial postnatal role of FOXG1 in neuronal maturation and myelination and elucidated the essential pathophysiology mechanisms of FS.
Figures




References
-
- Xuan S. et al. Winged helix transcription factor BF-1 is essential for the development of the cerebral hemispheres. Neuron 14, 1141–1152 (1995). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

