This is a preprint.
The variation and evolution of complete human centromeres
- PMID: 37398417
- PMCID: PMC10312506
- DOI: 10.1101/2023.05.30.542849
The variation and evolution of complete human centromeres
Update in
-
The variation and evolution of complete human centromeres.Nature. 2024 May;629(8010):136-145. doi: 10.1038/s41586-024-07278-3. Epub 2024 Apr 3. Nature. 2024. PMID: 38570684 Free PMC article.
Abstract
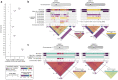
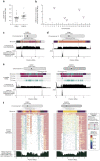
We completely sequenced and assembled all centromeres from a second human genome and used two reference sets to benchmark genetic, epigenetic, and evolutionary variation within centromeres from a diversity panel of humans and apes. We find that centromere single-nucleotide variation can increase by up to 4.1-fold relative to other genomic regions, with the caveat that up to 45.8% of centromeric sequence, on average, cannot be reliably aligned with current methods due to the emergence of new α-satellite higher-order repeat (HOR) structures and two to threefold differences in the length of the centromeres. The extent to which this occurs differs depending on the chromosome and haplotype. Comparing the two sets of complete human centromeres, we find that eight harbor distinctly different α-satellite HOR array structures and four contain novel α-satellite HOR variants in high abundance. DNA methylation and CENP-A chromatin immunoprecipitation experiments show that 26% of the centromeres differ in their kinetochore position by at least 500 kbp-a property not readily associated with novel α-satellite HORs. To understand evolutionary change, we selected six chromosomes and sequenced and assembled 31 orthologous centromeres from the common chimpanzee, orangutan, and macaque genomes. Comparative analyses reveal nearly complete turnover of α-satellite HORs, but with idiosyncratic changes in structure characteristic to each species. Phylogenetic reconstruction of human haplotypes supports limited to no recombination between the p- and q-arms of human chromosomes and reveals that novel α-satellite HORs share a monophyletic origin, providing a strategy to estimate the rate of saltatory amplification and mutation of human centromeric DNA.
Conflict of interest statement
COMPETING INTERESTS SN is now an employee of Oxford Nanopore Technologies, Inc.; SK has received travel funds to speak at events hosted by Oxford Nanopore Technologies, Inc.; EEE is a scientific advisory board member of Variant Bio, Inc.
Figures





References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
