This is a preprint.
The Genetic Architecture of Biological Age in Nine Human Organ Systems
- PMID: 37398441
- PMCID: PMC10312870
- DOI: 10.1101/2023.06.08.23291168
The Genetic Architecture of Biological Age in Nine Human Organ Systems
Update in
-
The genetic architecture of biological age in nine human organ systems.Nat Aging. 2024 Sep;4(9):1290-1307. doi: 10.1038/s43587-024-00662-8. Epub 2024 Jun 28. Nat Aging. 2024. PMID: 38942983 Free PMC article.
Abstract
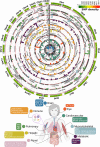
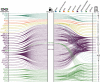
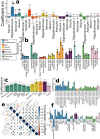
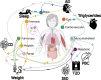
Understanding the genetic basis of biological aging in multi-organ systems is vital for elucidating age-related disease mechanisms and identifying therapeutic interventions. This study characterized the genetic architecture of the biological age gap (BAG) across nine human organ systems in 377,028 individuals of European ancestry from the UK Biobank. We discovered 393 genomic loci-BAG pairs (P-value<5×10-8) linked to the brain, eye, cardiovascular, hepatic, immune, metabolic, musculoskeletal, pulmonary, and renal systems. We observed BAG-organ specificity and inter-organ connections. Genetic variants associated with the nine BAGs are predominantly specific to the respective organ system while exerting pleiotropic effects on traits linked to multiple organ systems. A gene-drug-disease network confirmed the involvement of the metabolic BAG-associated genes in drugs targeting various metabolic disorders. Genetic correlation analyses supported Cheverud's Conjecture1 - the genetic correlation between BAGs mirrors their phenotypic correlation. A causal network revealed potential causal effects linking chronic diseases (e.g., Alzheimer's disease), body weight, and sleep duration to the BAG of multiple organ systems. Our findings shed light on promising therapeutic interventions to enhance human organ health within a complex multi-organ network, including lifestyle modifications and potential drug repositioning strategies for treating chronic diseases. All results are publicly available at https://labs-laboratory.com/medicine.
Conflict of interest statement
Competing Interests None
Figures


References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
