This is a preprint.
Single-cell sequencing of individual retinal organoids reveals determinants of cell fate heterogeneity
- PMID: 37398481
- PMCID: PMC10312535
- DOI: 10.1101/2023.05.31.543087
Single-cell sequencing of individual retinal organoids reveals determinants of cell fate heterogeneity
Update in
-
Single-cell sequencing of individual retinal organoids reveals determinants of cell-fate heterogeneity.Cell Rep Methods. 2023 Aug 9;3(8):100548. doi: 10.1016/j.crmeth.2023.100548. eCollection 2023 Aug 28. Cell Rep Methods. 2023. PMID: 37671011 Free PMC article.
Abstract
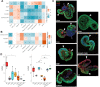
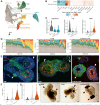
With a critical need for more complete in vitro models of human development and disease, organoids hold immense potential. Their complex cellular composition makes single-cell sequencing of great utility; however, the limitation of current technologies to a handful of treatment conditions restricts their use in screens or studies of organoid heterogeneity. Here, we apply sci-Plex, a single-cell combinatorial indexing (sci)-based RNA-seq multiplexing method to retinal organoids. We demonstrate that sci-Plex and 10x methods produce highly concordant cell class compositions and then expand sci-Plex to analyze the cell class composition of 410 organoids upon modulation of critical developmental pathways. Leveraging individual organoid data, we develop a method to measure organoid heterogeneity, and we identify that activation of Wnt signaling early in retinal organoid cultures increases retinal cell classes up to six weeks later. Our data show sci-Plex's potential to dramatically scale-up the analysis of treatment conditions on relevant human models.
Conflict of interest statement
Declaration of interests
C.T. is a SAB member, consultant and/or co-founder of Algen Biotechnologies, Altius Therapeutics, and Scale Biosciences. One or more embodiments of one or more patents and patent applications filed by the University of Washington may encompass methods, reagents, and the data disclosed in this manuscript. Some work in this study is related to technology described in patent applications.
Figures



References
-
- Eiraku M. et al. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature 472, 51–56 (2011). - PubMed
-
- Nakano T. et al. Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell 10, 771–785 (2012). - PubMed
-
- Xia Y. et al. Directed differentiation of human pluripotent cells to ureteric bud kidney progenitor-like cells. Nat. Cell Biol. 15, 1507–1515 (2013). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
