Overexpression of REDUCED WALL ACETYLATION C increases xylan acetylation and biomass recalcitrance in Populus
- PMID: 37399189
- PMCID: PMC10762510
- DOI: 10.1093/plphys/kiad377
Overexpression of REDUCED WALL ACETYLATION C increases xylan acetylation and biomass recalcitrance in Populus
Abstract
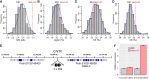
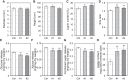
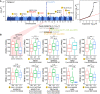
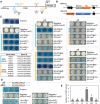
Plant lignocellulosic biomass, i.e. secondary cell walls of plants, is a vital alternative source for bioenergy. However, the acetylation of xylan in secondary cell walls impedes the conversion of biomass to biofuels. Previous studies have shown that REDUCED WALL ACETYLATION (RWA) proteins are directly involved in the acetylation of xylan but the regulatory mechanism of RWAs is not fully understood. In this study, we demonstrate that overexpression of a Populus trichocarpa PtRWA-C gene increases the level of xylan acetylation and increases the lignin content and S/G ratio, ultimately yielding poplar woody biomass with reduced saccharification efficiency. Furthermore, through gene coexpression network and expression quantitative trait loci (eQTL) analysis, we found that PtRWA-C was regulated not only by the secondary cell wall hierarchical regulatory network but also by an AP2 family transcription factor HARDY (HRD). Specifically, HRD activates PtRWA-C expression by directly binding to the PtRWA-C promoter, which is also the cis-eQTL for PtRWA-C. Taken together, our findings provide insights into the functional roles of PtRWA-C in xylan acetylation and consequently saccharification and shed light on synthetic biology approaches to manipulate this gene and alter cell wall properties. These findings have substantial implications for genetic engineering of woody species, which could be used as a sustainable source of biofuels, valuable biochemicals, and biomaterials.
© The Author(s) 2023. Published by Oxford University Press on behalf of American Society of Plant Biologists.
Conflict of interest statement
Conflict of interest statement. None declared.
Figures


References
-
- Busov V, Yordanov Y, Gou J, Meilan R, Ma C, Regan S, Strauss S. Activation tagging is an effective gene tagging system in Populus. Tree Genet Genomes. 2011:7:91–101. 10.1007/s11295-010-0317-7 - DOI
-
- Chen YK, Lin CH, Wang WC. The conversion of biomass into renewable jet fuel. Energy. 2020:201:117655. 10.1016/j.energy.2020.117655 - DOI
-
- Chow CN, Zheng HQ, Wu NY, Chien CH, Huang HD, Lee TY, Chiang-Hsieh YF, Hou PF, Yang TY, Chang WC. PlantPAN 2.0: an update of plant promoter analysis navigator for reconstructing transcriptional regulatory networks in plants. Nucleic Acids Res. 2016:44(D1):D1154–D1160. 10.1093/nar/gkv1035 - DOI - PMC - PubMed
-
- Ebringerová A. Structural diversity and application potential of hemicelluloses. Macromol Symp. 2005:232(1):1–12.

