Thousands of Pristionchus pacificus orphan genes were integrated into developmental networks that respond to diverse environmental microbiota
- PMID: 37399201
- PMCID: PMC10348561
- DOI: 10.1371/journal.pgen.1010832
Thousands of Pristionchus pacificus orphan genes were integrated into developmental networks that respond to diverse environmental microbiota
Abstract
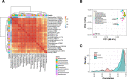
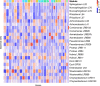
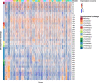
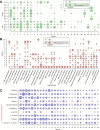
Adaptation of organisms to environmental change may be facilitated by the creation of new genes. New genes without homologs in other lineages are known as taxonomically-restricted orphan genes and may result from divergence or de novo formation. Previously, we have extensively characterized the evolution and origin of such orphan genes in the nematode model organism Pristionchus pacificus. Here, we employ large-scale transcriptomics to establish potential functional associations and to measure the degree of transcriptional plasticity among orphan genes. Specifically, we analyzed 24 RNA-seq samples from adult P. pacificus worms raised on 24 different monoxenic bacterial cultures. Based on coexpression analysis, we identified 28 large modules that harbor 3,727 diplogastrid-specific orphan genes and that respond dynamically to different bacteria. These coexpression modules have distinct regulatory architecture and also exhibit differential expression patterns across development suggesting a link between bacterial response networks and development. Phylostratigraphy revealed a considerably high number of family- and even species-specific orphan genes in certain coexpression modules. This suggests that new genes are not attached randomly to existing cellular networks and that integration can happen very fast. Integrative analysis of protein domains, gene expression and ortholog data facilitated the assignments of biological labels for 22 coexpression modules with one of the largest, fast-evolving module being associated with spermatogenesis. In summary, this work presents the first functional annotation for thousands of P. pacificus orphan genes and reveals insights into their integration into environmentally responsive gene networks.
Copyright: © 2023 Athanasouli et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
MeSH terms
LinkOut - more resources
Full Text Sources

