The NSL complex is required for piRNA production from telomeric clusters
- PMID: 37399316
- PMCID: PMC10313855
- DOI: 10.26508/lsa.202302194
The NSL complex is required for piRNA production from telomeric clusters
Abstract
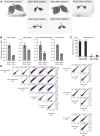
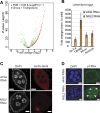
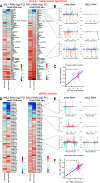
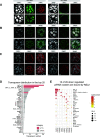
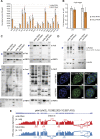
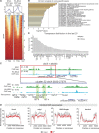
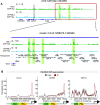
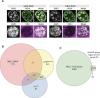
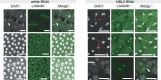
The NSL complex is a transcriptional activator. Germline-specific knockdown of NSL complex subunits NSL1, NSL2, and NSL3 results in reduced piRNA production from a subset of bidirectional piRNA clusters, accompanied by widespread transposon derepression. The piRNAs most transcriptionally affected by NSL2 and NSL1 RNAi map to telomeric piRNA clusters. At the chromatin level, these piRNA clusters also show decreased levels of H3K9me3, HP1a, and Rhino after NSL2 depletion. Using NSL2 ChIP-seq in ovaries, we found that this protein specifically binds promoters of telomeric transposons HeT-A, TAHRE, and TART Germline-specific depletion of NSL2 also led to a reduction in nuclear Piwi in nurse cells. Our findings thereby support a role for the NSL complex in promoting the transcription of piRNA precursors from telomeric piRNA clusters and in regulating Piwi levels in the Drosophila female germline.
© 2023 Iyer et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures







References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
