Highly Abundant Proteins Are Highly Thermostable
- PMID: 37399326
- PMCID: PMC10317291
- DOI: 10.1093/gbe/evad112
Highly Abundant Proteins Are Highly Thermostable
Abstract
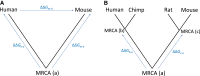
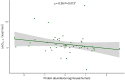
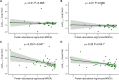
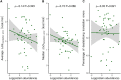
Highly abundant proteins tend to evolve slowly (a trend called E-R anticorrelation), and a number of hypotheses have been proposed to explain this phenomenon. The misfolding avoidance hypothesis attributes the E-R anticorrelation to the abundance-dependent toxic effects of protein misfolding. To avoid these toxic effects, protein sequences (particularly those of highly expressed proteins) would be under selection to fold properly. One prediction of the misfolding avoidance hypothesis is that highly abundant proteins should exhibit high thermostability (i.e., a highly negative free energy of folding, ΔG). Thus far, only a handful of analyses have tested for a relationship between protein abundance and thermostability, producing contradictory results. These analyses have been limited by 1) the scarcity of ΔG data, 2) the fact that these data have been obtained by different laboratories and under different experimental conditions, 3) the problems associated with using proteins' melting energy (Tm) as a proxy for ΔG, and 4) the difficulty of controlling for potentially confounding variables. Here, we use computational methods to compare the free energy of folding of pairs of human-mouse orthologous proteins with different expression levels. Even though the effect size is limited, the most highly expressed ortholog is often the one with a more negative ΔG of folding, indicating that highly expressed proteins are often more thermostable.
Keywords: expression levels; misfolding avoidance hypothesis; protein thermostability; rates of evolution; translational robustness hypothesis.
© The Author(s) 2023. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures
References
-
- Alvarez-Ponce D. 2014. Why proteins evolve at different rates: the determinants of proteins’ rates of evolution. In: Fares MA, editor. Natural selection: methods and applications. London: CRC Press (Taylor & Francis). p. 126–178.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

