Phase variation as a major mechanism of adaptation in Mycobacterium tuberculosis complex
- PMID: 37399390
- PMCID: PMC10334774
- DOI: 10.1073/pnas.2301394120
Phase variation as a major mechanism of adaptation in Mycobacterium tuberculosis complex
Abstract
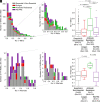
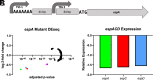
Phase variation induced by insertions and deletions (INDELs) in genomic homopolymeric tracts (HT) can silence and regulate genes in pathogenic bacteria, but this process is not characterized in MTBC (Mycobacterium tuberculosis complex) adaptation. We leverage 31,428 diverse clinical isolates to identify genomic regions including phase-variants under positive selection. Of 87,651 INDEL events that emerge repeatedly across the phylogeny, 12.4% are phase-variants within HTs (0.02% of the genome by length). We estimated the in-vitro frameshift rate in a neutral HT at 100× the neutral substitution rate at [Formula: see text] frameshifts/HT/year. Using neutral evolution simulations, we identified 4,098 substitutions and 45 phase-variants to be putatively adaptive to MTBC (P < 0.002). We experimentally confirm that a putatively adaptive phase-variant alters the expression of espA, a critical mediator of ESX-1-dependent virulence. Our evidence supports the hypothesis that phase variation in the ESX-1 system of MTBC can act as a toggle between antigenicity and survival in the host.
Keywords: genomics; microbiology; phase variation; tuberculosis.
Conflict of interest statement
The authors declare no competing interest.
Figures


References
-
- Shaw G. B., “Practical uses of litmus paper in Möbius strips” (Tech. Rep. CUCS-29-82, Columbia University, NY, 1982).
-
- Gagneux S., Ecology and evolution of Mycobacterium tuberculosis. Nat. Rev. Microbiol. 16, 202–213 (2018). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

