Loss of the batten disease protein CLN3 leads to mis-trafficking of M6PR and defective autophagic-lysosomal reformation
- PMID: 37400440
- PMCID: PMC10317969
- DOI: 10.1038/s41467-023-39643-7
Loss of the batten disease protein CLN3 leads to mis-trafficking of M6PR and defective autophagic-lysosomal reformation
Abstract
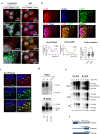
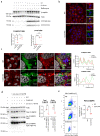
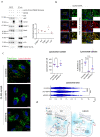
Batten disease, one of the most devastating types of neurodegenerative lysosomal storage disorders, is caused by mutations in CLN3. Here, we show that CLN3 is a vesicular trafficking hub connecting the Golgi and lysosome compartments. Proteomic analysis reveals that CLN3 interacts with several endo-lysosomal trafficking proteins, including the cation-independent mannose 6 phosphate receptor (CI-M6PR), which coordinates the targeting of lysosomal enzymes to lysosomes. CLN3 depletion results in mis-trafficking of CI-M6PR, mis-sorting of lysosomal enzymes, and defective autophagic lysosomal reformation. Conversely, CLN3 overexpression promotes the formation of multiple lysosomal tubules, which are autophagy and CI-M6PR-dependent, generating newly formed proto-lysosomes. Together, our findings reveal that CLN3 functions as a link between the M6P-dependent trafficking of lysosomal enzymes and lysosomal reformation pathway, explaining the global impairment of lysosomal function in Batten disease.
© 2023. The Author(s).
Conflict of interest statement
A.B. is co-founder of CASMA Therapeutics and advisory board member of Next Generation Diagnostics and Avilar and Coave Therapeutics. The remaining authors declare no competing interests.
Figures





References
-
- Bennett, M. J. & Hofmann, S. L. The neuronal ceroid-lipofuscinoses (Batten disease): A new class of lysosomal storage diseases. in Journal of Inherited Metabolic Disease vol. 22 535–544 (J Inherit Metab Dis, 1999). 10.1023/A:1005564509027. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

