A catalog of bacterial reference genomes from cultivated human oral bacteria
- PMID: 37400465
- PMCID: PMC10318035
- DOI: 10.1038/s41522-023-00414-3
A catalog of bacterial reference genomes from cultivated human oral bacteria
Abstract
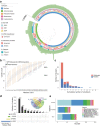
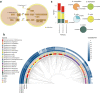
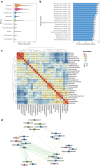
The oral cavity harbors highly diverse communities of microorganisms. However, the number of isolated species and high-quality genomes is limited. Here we present a Cultivated Oral Bacteria Genome Reference (COGR), comprising 1089 high-quality genomes based on large-scale aerobic and anaerobic cultivation of human oral bacteria isolated from dental plaques, tongue, and saliva. COGR covers five phyla and contains 195 species-level clusters of which 95 include 315 genomes representing species with no taxonomic annotation. The oral microbiota differs markedly between individuals, with 111 clusters being person-specific. Genes encoding CAZymes are abundant in the genomes of COGR. Members of the Streptococcus genus make up the largest proportion of COGR and many of these harbor entire pathways for quorum sensing important for biofilm formation. Several clusters containing unknown bacteria are enriched in individuals with rheumatoid arthritis, emphasizing the importance of culture-based isolation for characterizing and exploiting oral bacteria.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

