Arf1 coordinates fatty acid metabolism and mitochondrial homeostasis
- PMID: 37400497
- PMCID: PMC10415182
- DOI: 10.1038/s41556-023-01180-2
Arf1 coordinates fatty acid metabolism and mitochondrial homeostasis
Abstract
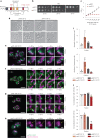
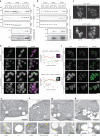
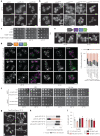
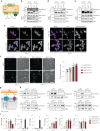
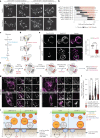
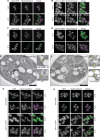
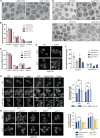
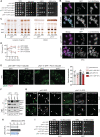
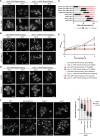
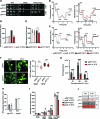
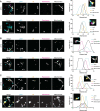
Lipid mobilization through fatty acid β-oxidation is a central process essential for energy production during nutrient shortage. In yeast, this catabolic process starts in the peroxisome from where β-oxidation products enter mitochondria and fuel the tricarboxylic acid cycle. Little is known about the physical and metabolic cooperation between these organelles. Here we found that expression of fatty acid transporters and of the rate-limiting enzyme involved in β-oxidation is decreased in cells expressing a hyperactive mutant of the small GTPase Arf1, leading to an accumulation of fatty acids in lipid droplets. Consequently, mitochondria became fragmented and ATP synthesis decreased. Genetic and pharmacological depletion of fatty acids phenocopied the arf1 mutant mitochondrial phenotype. Although β-oxidation occurs in both mitochondria and peroxisomes in mammals, Arf1's role in fatty acid metabolism is conserved. Together, our results indicate that Arf1 integrates metabolism into energy production by regulating fatty acid storage and utilization, and presumably organelle contact sites.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures







References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

