Identification of cuproptosis-related lncRNA for predicting prognosis and immunotherapeutic response in cervical cancer
- PMID: 37400520
- PMCID: PMC10318051
- DOI: 10.1038/s41598-023-37898-0
Identification of cuproptosis-related lncRNA for predicting prognosis and immunotherapeutic response in cervical cancer
Abstract
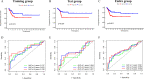
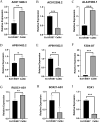
Patients diagnosed with advanced cervical cancer (CC) have poor prognosis after primary treatment, and there is a lack of biomarkers for predicting patients with an increased risk of recurrence of CC. Cuproptosis is reported to play a role in tumorigenesis and progression. However, the clinical impacts of cuproptosis-related lncRNAs (CRLs) in CC remain largely unclear. Our study attempted to identify new potential biomarkers to predict prognosis and response to immunotherapy with the aim of improving this situation. The transcriptome data, MAF files, and clinical information for CC cases were obtained from the cancer genome atlas, and Pearson correlation analysis was utilized to identify CRLs. In total, 304 eligible patients with CC were randomly assigned to training and test groups. LASSO regression and multivariate Cox regression were performed to construct a cervical cancer prognostic signature based on cuproptosis-related lncRNAs. Afterwards, we generated Kaplan-Meier curves, receiver operating characteristic curves and nomograms to verify the ability to predict prognosis of patients with CC. Genes for assessing differential expression among risk subgroups were also evaluated by functional enrichment analysis. Immune cell infiltration and the tumour mutation burden were analysed to explore the underlying mechanisms of the signature. Furthermore, the potential value of the prognostic signature to predict response to immunotherapy and sensitivity to chemotherapy drugs was examined. In our study, a risk signature containing eight cuproptosis-related lncRNAs (AL441992.1, SOX21-AS1, AC011468.3, AC012306.2, FZD4-DT, AP001922.5, RUSC1-AS1, AP001453.2) to predict the survival outcome of CC patients was developed, and the reliability of the risk signature was appraised. Cox regression analyses indicated that the comprehensive risk score is an independent prognostic factor. Moreover, significant differences were found in progression-free survival, immune cell infiltration, therapeutic response to immune checkpoint inhibitors, and IC50 for chemotherapeutic agents between risk subgroups, suggesting that our model can be well employed to assess the clinical efficacy of immunotherapy and chemotherapy. Based on our 8-CRLs risk signature, we were able to independently assess the outcome and response to immunotherapy of CC patients, and this signature might benefit clinical decision-making for individualized treatment.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures









Similar articles
-
Prognostic signature construction and immunotherapy response analysis for Uterine Corpus Endometrial Carcinoma based on cuproptosis-related lncRNAs.Comput Biol Med. 2023 Jun;159:106905. doi: 10.1016/j.compbiomed.2023.106905. Epub 2023 Apr 11. Comput Biol Med. 2023. PMID: 37060773
-
Signature of seven cuproptosis-related lncRNAs as a novel biomarker to predict prognosis and therapeutic response in cervical cancer.Front Genet. 2022 Sep 20;13:989646. doi: 10.3389/fgene.2022.989646. eCollection 2022. Front Genet. 2022. PMID: 36204323 Free PMC article.
-
Development and validation of cuproptosis-related lncRNA signatures for prognosis prediction in colorectal cancer.BMC Med Genomics. 2023 Mar 22;16(1):58. doi: 10.1186/s12920-023-01487-x. BMC Med Genomics. 2023. PMID: 36949429 Free PMC article.
-
A novel prognostic signature for lung adenocarcinoma based on cuproptosis-related lncRNAs: A Review.Medicine (Baltimore). 2022 Dec 9;101(49):e31924. doi: 10.1097/MD.0000000000031924. Medicine (Baltimore). 2022. PMID: 36626411 Free PMC article. Review.
-
Review to Elucidate the Correlation between Cuproptosis-Related Genes and Immune Infiltration for Enhancing the Detection and Treatment of Cervical Cancer.Int J Mol Sci. 2024 Oct 1;25(19):10604. doi: 10.3390/ijms251910604. Int J Mol Sci. 2024. PMID: 39408933 Free PMC article. Review.
Cited by
-
Integrating Clinical and Transcriptomic Profiles Associated with Vitamin D to Enhance Disease-Free Survival in Cervical Cancer Recurrence Using the CatBoost Algorithm.Diagnostics (Basel). 2025 Jun 21;15(13):1579. doi: 10.3390/diagnostics15131579. Diagnostics (Basel). 2025. PMID: 40647578 Free PMC article.
-
Noncoding RNAs as an emerging resistance mechanism to immunotherapies in cancer: basic evidence and therapeutic implications.Front Immunol. 2023 Sep 12;14:1268745. doi: 10.3389/fimmu.2023.1268745. eCollection 2023. Front Immunol. 2023. PMID: 37767098 Free PMC article. Review.
-
Exploring program-cell death patterns to predict prognosis and sensitivity of cervical cancer immunotherapy via multi-omics analysis and clinical samples.Discov Oncol. 2025 May 28;16(1):940. doi: 10.1007/s12672-025-02622-z. Discov Oncol. 2025. PMID: 40434537 Free PMC article.
-
Why Is Surgery Still Done after Concurrent Chemoradiotherapy in Locally Advanced Cervical Cancer in Romania?Cancers (Basel). 2024 Jan 19;16(2):425. doi: 10.3390/cancers16020425. Cancers (Basel). 2024. PMID: 38275866 Free PMC article.
-
SRY-box transcription factor 21 antisense divergent transcript 1: Regulatory roles and clinical significance in neoplastic conditions and Alzheimer's Disease.J Cancer. 2023 Oct 2;14(17):3258-3274. doi: 10.7150/jca.89619. eCollection 2023. J Cancer. 2023. PMID: 37928430 Free PMC article. Review.

