Spatial mapping of cellular senescence: emerging challenges and opportunities
- PMID: 37400722
- PMCID: PMC10505496
- DOI: 10.1038/s43587-023-00446-6
Spatial mapping of cellular senescence: emerging challenges and opportunities
Abstract
Cellular senescence is a well-established driver of aging and age-related diseases. There are many challenges to mapping senescent cells in tissues such as the absence of specific markers and their relatively low abundance and vast heterogeneity. Single-cell technologies have allowed unprecedented characterization of senescence; however, many methodologies fail to provide spatial insights. The spatial component is essential, as senescent cells communicate with neighboring cells, impacting their function and the composition of extracellular space. The Cellular Senescence Network (SenNet), a National Institutes of Health (NIH) Common Fund initiative, aims to map senescent cells across the lifespan of humans and mice. Here, we provide a comprehensive review of the existing and emerging methodologies for spatial imaging and their application toward mapping senescent cells. Moreover, we discuss the limitations and challenges inherent to each technology. We argue that the development of spatially resolved methods is essential toward the goal of attaining an atlas of senescent cells.
© 2023. Springer Nature America, Inc.
Conflict of interest statement
Competing interests
J.H.L. is an inventor on pending patent applications related to Seq-Scope. All other authors declare no competing interests.
Figures

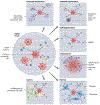



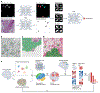
References
Publication types
MeSH terms
Grants and funding
- U54 AG076041/AG/NIA NIH HHS/United States
- U54 AG076040/AG/NIA NIH HHS/United States
- UH3 CA268091/CA/NCI NIH HHS/United States
- UG3 CA268091/CA/NCI NIH HHS/United States
- U54 AG075932/AG/NIA NIH HHS/United States
- U54 AG075931/AG/NIA NIH HHS/United States
- U54 AG079758/AG/NIA NIH HHS/United States
- UG3 CA268103/CA/NCI NIH HHS/United States
- U54 AG075941/AG/NIA NIH HHS/United States
- UG3 CA268096/CA/NCI NIH HHS/United States
- U54 AG075936/AG/NIA NIH HHS/United States
- UG3 CA268112/CA/NCI NIH HHS/United States
- UH3 CA268103/CA/NCI NIH HHS/United States
- UG3 CA268202/CA/NCI NIH HHS/United States
- U24 CA268108/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical

