Krüppel-like factor 7 deficiency disrupts corpus callosum development and neuronal migration in the developing mouse cerebral cortex
- PMID: 37401095
- PMCID: PMC10467035
- DOI: 10.1111/bpa.13186
Krüppel-like factor 7 deficiency disrupts corpus callosum development and neuronal migration in the developing mouse cerebral cortex
Abstract
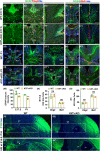
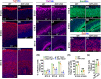
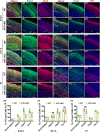
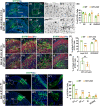
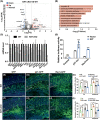
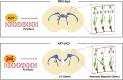
Krüppel-like Factor 7 (KLF7) is a zinc finger transcription factor that has a critical role in cellular differentiation, tumorigenesis, and regeneration. Mutations in Klf7 are associated with autism spectrum disorder, which is characterized by neurodevelopmental delay and intellectual disability. Here we show that KLF7 regulates neurogenesis and neuronal migration during mouse cortical development. Conditional depletion of KLF7 in neural progenitor cells resulted in agenesis of the corpus callosum, defects in neurogenesis, and impaired neuronal migration in the neocortex. Transcriptomic profiling analysis indicated that KLF7 regulates a cohort of genes involved in neuronal differentiation and migration, including p21 and Rac3. These findings provide insights into our understanding of the potential mechanisms underlying neurological defects associated with Klf7 mutations.
Keywords: corpus callosum; krüppel-like factor 7; neocortex; neurogenesis; neuronal migration.
© 2023 The Authors. Brain Pathology published by John Wiley & Sons Ltd on behalf of International Society of Neuropathology.
Conflict of interest statement
The authors declare no competing or financial interests.
Figures

References
-
- Martinez‐Cerdeno V, Noctor SC, Kriegstein AR. The role of intermediate progenitor cells in the evolutionary expansion of the cerebral cortex. Cereb Cortex. 2006;16(Suppl 1):i152–61. - PubMed
-
- Angevine JB Jr, Sidman RL. Autoradiographic study of cell migration during histogenesis of cerebral cortex in the mouse. Nature. 1961;192:766–8. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

