Computational design of matrix metalloprotenaise-9 (MMP-9) resistant to auto-cleavage
- PMID: 37401540
- PMCID: PMC10422929
- DOI: 10.1042/BCJ20230139
Computational design of matrix metalloprotenaise-9 (MMP-9) resistant to auto-cleavage
Abstract
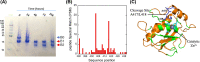
Matrix metalloproteinase-9 (MMP-9) is an endopeptidase that remodels the extracellular matrix. MMP-9 has been implicated in several diseases including neurodegeneration, arthritis, cardiovascular diseases, fibrosis and several types of cancer, resulting in a high demand for MMP-9 inhibitors for therapeutic purposes. For such drug design efforts, large amounts of MMP-9 are required. Yet, the catalytic domain of MMP-9 (MMP-9Cat) is an intrinsically unstable enzyme that tends to auto-cleave within minutes, making it difficult to use in drug design experiments and other biophysical studies. We set our goal to design MMP-9Cat variant that is active but stable to auto-cleavage. For this purpose, we first identified potential auto-cleavage sites on MMP-9Cat using mass spectroscopy and then eliminated the auto-cleavage site by predicting mutations that minimize auto-cleavage potential without reducing enzyme stability. Four computationally designed MMP-9Cat variants were experimentally constructed and evaluated for auto-cleavage and enzyme activity. Our best variant, Des2, with 2 mutations, was as active as the wild-type enzyme but did not exhibit auto-cleavage after 7 days of incubation at 37°C. This MMP-9Cat variant, with an identical with MMP-9Cat WT active site, is an ideal candidate for drug design experiments targeting MMP-9 and enzyme crystallization experiments. The developed strategy for MMP-9CAT stabilization could be applied to redesign other proteases to improve their stability for various biotechnological applications.
Keywords: MMP-9; auto-cleavage; auto-degradation; enzyme engineering; matrix metalloproteases; protein engineering.
© 2023 The Author(s).
Conflict of interest statement
The authors declare that there are no competing interests associated with the manuscript.
Figures


Update of
-
Computational design of Matrix Metalloprotenaise-9 (MMP-9) resistant to auto-cleavage.bioRxiv [Preprint]. 2023 Apr 11:2023.04.11.536383. doi: 10.1101/2023.04.11.536383. bioRxiv. 2023. Update in: Biochem J. 2023 Jul 26;480(14):1097-1107. doi: 10.1042/BCJ20230139. PMID: 37090502 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

