Breast cancer PDxO cultures for drug discovery and functional precision oncology
- PMID: 37402170
- PMCID: PMC10339058
- DOI: 10.1016/j.xpro.2023.102402
Breast cancer PDxO cultures for drug discovery and functional precision oncology
Erratum in
-
Breast cancer PDxO cultures for drug discovery and functional precision oncology.STAR Protoc. 2025 Mar 21;6(1):103676. doi: 10.1016/j.xpro.2025.103676. Epub 2025 Feb 21. STAR Protoc. 2025. PMID: 39985776 Free PMC article. No abstract available.
Abstract
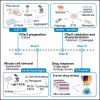
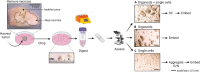
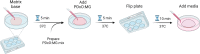
Patient-derived xenografts (PDXs) have clinical value but are time-, cost-, and labor-intensive and thus ill-suited for large-scale experiments. Here, we present a protocol to convert PDX tumors into PDxOs for long-term cultures amenable to moderate-throughput drug screens, including in-depth PDxO validation. We describe steps for PDxO preparation and mouse cell removal. We then detail PDxO validation and characterization and drug response assay. Our PDxO drug screening platform can predict therapy response in vivo and inform functional precision oncology for patients. For complete details on the use and execution of this protocol, please refer to Guillen et al.1.
Keywords: Cancer; Cell Culture; Cell-based Assays; Model Organisms; Organoids.
Copyright © 2023 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The University of Utah may license the PDX and PDxO models described here to for-profit companies, which may result in tangible property royalties to members of the Welm lab who developed the models (S.D.S., A.J.B., L.Z., C.H.Y., E.C.S., K.P.G., B.E.W., and A.L.W.).
Figures







References
-
- Guillen K.P., Fujita M., Butterfield A.J., Scherer S.D., Bailey M.H., Chu Z., DeRose Y.S., Zhao L., Cortes-Sanchez E., Yang C.-H., et al. A human breast cancer-derived xenograft and organoid platform for drug discovery and presicion oncology. Nat. Cancer. 2022;3:232–250. doi: 10.1038/s43018-022-00337-6. - DOI - PMC - PubMed
-
- DeRose Y.S., Gligorich K.M., Wang G., Georgelas A., Bowman P., Courdy S.J., Welm A.L., Welm B.E. Patient-derived models of human breast cancer: protocols for in vitro and in vivo applications in tumor biology and translational medicine. Curr. Protoc. Pharmacol. 2013;Chapter 14:Unit14.23. doi: 10.1002/0471141755.ph1423s60. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

