Transcription factors interact with RNA to regulate genes
- PMID: 37402367
- PMCID: PMC10529847
- DOI: 10.1016/j.molcel.2023.06.012
Transcription factors interact with RNA to regulate genes
Abstract
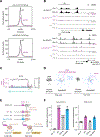
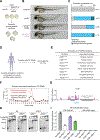
Transcription factors (TFs) orchestrate the gene expression programs that define each cell's identity. The canonical TF accomplishes this with two domains, one that binds specific DNA sequences and the other that binds protein coactivators or corepressors. We find that at least half of TFs also bind RNA, doing so through a previously unrecognized domain with sequence and functional features analogous to the arginine-rich motif of the HIV transcriptional activator Tat. RNA binding contributes to TF function by promoting the dynamic association between DNA, RNA, and TF on chromatin. TF-RNA interactions are a conserved feature important for vertebrate development and disrupted in disease. We propose that the ability to bind DNA, RNA, and protein is a general property of many TFs and is fundamental to their gene regulatory function.
Keywords: RNA; RNA-binding proteins; arginine-rich motif; chromatin; development; gene regulation; single-molecule imaging; transcription factor; zebrafish.
Copyright © 2023 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests R.A.Y. is a founder and shareholder of Syros Pharmaceuticals, Camp4 Therapeutics, Omega Therapeutics, Dewpoint Therapeutics, and Paratus Sciences. R.A.Y. is a member of Molecular Cell’s advisory board. O.O. and J.E.H. are consultants at Camp4 Therapeutics. L.I.Z. is a founder and stockholder of Fate Therapeutics, Camp4 Therapeutics, Amagma Therapeutics, Scholar Rock, and Branch Biosciences. L.I.Z. is a consultant for Celularity and Cellarity. The Whitehead Institute has filed a patent application related to this work.
Figures





Comment in
-
RNA binding - a useful third wheel in transcription.Nat Rev Mol Cell Biol. 2023 Sep;24(9):604. doi: 10.1038/s41580-023-00643-6. Nat Rev Mol Cell Biol. 2023. PMID: 37474726 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

