Identification of 2-Sulfonyl/Sulfonamide Pyrimidines as Covalent Inhibitors of WRN Using a Multiplexed High-Throughput Screening Assay
- PMID: 37403936
- PMCID: PMC10358344
- DOI: 10.1021/acs.biochem.2c00599
Identification of 2-Sulfonyl/Sulfonamide Pyrimidines as Covalent Inhibitors of WRN Using a Multiplexed High-Throughput Screening Assay
Abstract
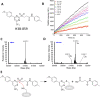
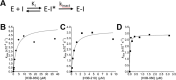
Werner syndrome protein (WRN) is a multifunctional enzyme with helicase, ATPase, and exonuclease activities that are necessary for numerous DNA-related transactions in the human cell. Recent studies identified WRN as a synthetic lethal target in cancers characterized by genomic microsatellite instability resulting from defects in DNA mismatch repair pathways. WRN's helicase activity is essential for the viability of these high microsatellite instability (MSI-H) cancers and thus presents a therapeutic opportunity. To this end, we developed a multiplexed high-throughput screening assay that monitors exonuclease, ATPase, and helicase activities of full-length WRN. This screening campaign led to the discovery of 2-sulfonyl/sulfonamide pyrimidine derivatives as novel covalent inhibitors of WRN helicase activity. The compounds are specific for WRN versus other human RecQ family members and show competitive behavior with ATP. Examination of these novel chemical probes established the sulfonamide NH group as a key driver of compound potency. One of the leading compounds, H3B-960, showed consistent activities in a range of assays (IC50 = 22 nM, KD = 40 nM, KI = 32 nM), and the most potent compound identified, H3B-968, has inhibitory activity IC50 ∼ 10 nM. These kinetic properties trend toward other known covalent druglike molecules. Our work provides a new avenue for screening WRN for inhibitors that may be adaptable to different therapeutic modalities such as targeted protein degradation, as well as a proof of concept for the inhibition of WRN helicase activity by covalent molecules.
Conflict of interest statement
The authors declare the following competing financial interest(s): The authors are former employees of H3 Biomedicine.
Figures




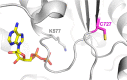
References
-
- Larsen N. B.; Hickson I. D.. RecQ Helicases: Conserved Guardians of Genomic Integrity. In Advances in Experimental Medicine and Biology, 2013; Vol. 767, pp 161–184. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

