A novel Gardnerella, Prevotella, and Lactobacillus standard that improves accuracy in quantifying bacterial burden in vaginal microbial communities
- PMID: 37404722
- PMCID: PMC10315654
- DOI: 10.3389/fcimb.2023.1198113
A novel Gardnerella, Prevotella, and Lactobacillus standard that improves accuracy in quantifying bacterial burden in vaginal microbial communities
Abstract
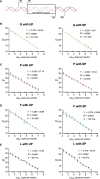
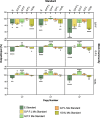
Bacterial vaginosis (BV) is the most common vaginal dysbiosis. In this condition, a polymicrobial biofilm develops on vaginal epithelial cells. Accurately quantifying the bacterial burden of the BV biofilm is necessary to further our understanding of BV pathogenesis. Historically, the standard for calculating total bacterial burden of the BV biofilm has been based on quantifying Escherichia coli 16S rRNA gene copy number. However, E. coli is improper for measuring the bacterial burden of this unique micro-environment. Here, we propose a novel qPCR standard to quantify bacterial burden in vaginal microbial communities, from an optimal state to a mature BV biofilm. These standards consist of different combinations of vaginal bacteria including three common BV-associated bacteria (BVAB) Gardnerella spp. (G), Prevotella spp. (P), and Fannyhessea spp. (F) and commensal Lactobacillus spp. (L) using the 16S rRNA gene (G:P:F:L, G:P:F, G:P:L and 1G:9L). We compared these standards to the traditional E. coli (E) reference standard using known quantities of mock vaginal communities and 16 vaginal samples from women. The E standard significantly underestimated the copy numbers of the mock communities, and this underestimation was significantly greater at lower copy numbers of these communities. The G:P:L standard was the most accurate across all mock communities and when compared to other mixed vaginal standards. Mixed vaginal standards were further validated with vaginal samples. This new G:P:L standard can be used in BV pathogenesis research to enhance reproducibility and reliability in quantitative measurements of BVAB, spanning from the optimal to non-optimal (including BV) vaginal microbiota.
Keywords: Gardnerella; Lactobacillus; Prevotella; bacterial burden; bacterial vaginosis; biofilm; qPCR standard; vaginal microbiome.
Copyright © 2023 Elnaggar, Ardizzone, Cerca, Toh, Łaniewski, Lillis, Herbst-Kralovetz, Quayle, Muzny and Taylor.
Conflict of interest statement
Author CM is a consultant for Scynexis, Cepheid, and BioNTech. Author CM has received research funding support from NIH/NIAID, Lupin Pharmaceuticals, Abbott Molecular, Visby, and Gilead as well as honoraria from Visby Medical, Elsevier, Abbott Molecular, Scynexis, Roche, and Cepheid. Author RL is the clinical trial site PI for Cepheid, Hologic, Gilead, Visby, Roche, Becton Dickenson, OrthoQuidel, Merck, BioFire, CUE, CovarsaDx, and a consultant for Roche and Cepheid. Author MH-K is on the scientific advisory board for Freya Biosciences and a consultant for Vaginal Biome Sciences. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

