Immunogenicity and efficacy of a novel multi-patch SARS-CoV-2/COVID-19 vaccine candidate
- PMID: 37404819
- PMCID: PMC10316789
- DOI: 10.3389/fimmu.2023.1160065
Immunogenicity and efficacy of a novel multi-patch SARS-CoV-2/COVID-19 vaccine candidate
Abstract
Introduction: While there has been considerable progress in the development of vaccines against SARS-CoV-2, largely based on the S (spike) protein of the virus, less progress has been made with vaccines delivering different viral antigens with cross-reactive potential.
Methods: In an effort to develop an immunogen with the capacity to induce broad antigen presentation, we have designed a multi-patch synthetic candidate containing dominant and persistent B cell epitopes from conserved regions of SARS-CoV-2 structural proteins associated with long-term immunity, termed CoV2-BMEP. Here we describe the characterization, immunogenicity and efficacy of CoV2-BMEP using two delivery platforms: nucleic acid DNA and attenuated modified vaccinia virus Ankara (MVA).
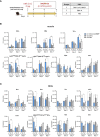
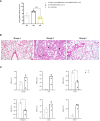
Results: In cultured cells, both vectors produced a main protein of about 37 kDa as well as heterogeneous proteins with size ranging between 25-37 kDa. In C57BL/6 mice, both homologous and heterologous prime/boost combination of vectors induced the activation of SARS-CoV-2-specific CD4 and CD8 T cell responses, with a more balanced CD8+ T cell response detected in lungs. The homologous MVA/MVA immunization regimen elicited the highest specific CD8+ T cell responses in spleen and detectable binding antibodies (bAbs) to S and N antigens of SARS-CoV-2. In SARS-CoV-2 susceptible k18-hACE2 Tg mice, two doses of MVA-CoV2-BMEP elicited S- and N-specific bAbs as well as cross-neutralizing antibodies against different variants of concern (VoC). After SARS-CoV-2 challenge, all animals in the control unvaccinated group succumbed to the infection while vaccinated animals with high titers of neutralizing antibodies were fully protected against mortality, correlating with a reduction of virus infection in the lungs and inhibition of the cytokine storm.
Discussion: These findings revealed a novel immunogen with the capacity to control SARS-CoV-2 infection, using a broader antigen presentation mechanism than the approved vaccines based solely on the S antigen.
Keywords: B and T cell immune responses; SARS-CoV-2; binding and neutralizing antibodies; efficacy; mice studies; multi-patch vaccine; poxvirus MVA.
Copyright © 2023 Perdiguero, Marcos-Villar, López-Bravo, Sánchez-Cordón, Zamora, Valverde, Sorzano, Sin, Álvarez, Ramos, Del Val, Esteban and Gómez.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures







References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

