Crystal structure of CmnB involved in the biosynthesis of the nonproteinogenic amino acid L-2,3-diaminopropionic acid
- PMID: 37405487
- PMCID: PMC10327575
- DOI: 10.1107/S2053230X23005769
Crystal structure of CmnB involved in the biosynthesis of the nonproteinogenic amino acid L-2,3-diaminopropionic acid
Abstract
L-2,3-Diaminopropionic acid (L-Dap) is a nonproteinogenic amino acid that plays as an important role as a building block in the biosynthesis of several natural products, including capreomycin, viomycin, zwittermicin, staphyloferrin and dapdiamide. A previous study reported that CmnB and CmnK are two enzymes that are involved in the formation of L-Dap in the biosynthesis of capreomycin. CmnB catalyzes the condensation reaction of O-phospho-L-serine and L-glutamic acid to generate N-(1-amino-1-carboxyl-2-ethyl)glutamic acid, which subsequently undergoes oxidative hydrolysis via CmnK to generate the product L-Dap. Here, the crystal structure of CmnB in complex with the reaction intermediate PLP-α-aminoacrylate is reported at 2.2 Å resolution. Notably, CmnB is the second known example of a PLP-dependent enzyme that forms a monomeric structure in crystal packing. The crystal structure of CmnB also provides insights into the catalytic mechanism of the enzyme and supports the biosynthetic pathway of L-Dap reported in previous studies.
Keywords: CmnB; PLP-dependent enzymes; capreomycin biosynthesis; l-2,3-diaminopropionic acid; l-Dap.
Figures



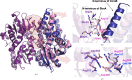
References
-
- Agirre, J., Atanasova, M., Bagdonas, H., Ballard, C. B., Baslé, A., Beilsten-Edmands, J., Borges, R. J., Brown, D. G., Burgos-Mármol, J. J., Berrisford, J. M., Bond, P. S., Caballero, I., Catapano, L., Chojnowski, G., Cook, A. G., Cowtan, K. D., Croll, T. I., Debreczeni, J. É., Devenish, N. E., Dodson, E. J., Drevon, T. R., Emsley, P., Evans, G., Evans, P. R., Fando, M., Foadi, J., Fuentes-Montero, L., Garman, E. F., Gerstel, M., Gildea, R. J., Hatti, K., Hekkelman, M. L., Heuser, P., Hoh, S. W., Hough, M. A., Jenkins, H. T., Jiménez, E., Joosten, R. P., Keegan, R. M., Keep, N., Krissinel, E. B., Kolenko, P., Kovalevskiy, O., Lamzin, V. S., Lawson, D. M., Lebedev, A. A., Leslie, A. G. W., Lohkamp, B., Long, F., Malý, M., McCoy, A. J., McNicholas, S. J., Medina, A., Millán, C., Murray, J. W., Murshudov, G. N., Nicholls, R. A., Noble, M. E. M., Oeffner, R., Pannu, N. S., Parkhurst, J. M., Pearce, N., Pereira, J., Perrakis, A., Powell, H. R., Read, R. J., Rigden, D. J., Rochira, W., Sammito, M., Sánchez Rodríguez, F., Sheldrick, G. M., Shelley, K. L., Simkovic, F., Simpkin, A. J., Skubak, P., Sobolev, E., Steiner, R. A., Stevenson, K., Tews, I., Thomas, J. M. H., Thorn, A., Valls, J. T., Uski, V., Usón, I., Vagin, A., Velankar, S., Vollmar, M., Walden, H., Waterman, D., Wilson, K. S., Winn, M. D., Winter, G., Wojdyr, M. & Yamashita, K. (2023). Acta Cryst. D79, 449–461.
-
- Chang, C.-Y., Lyu, S.-Y., Liu, Y.-C., Hsu, N.-S., Wu, C.-C., Tang, C.-F., Lin, K.-H., Ho, J.-Y., Wu, C.-J., Tsai, M.-D. & Li, T.-L. (2014). Angew. Chem. Int. Ed. 53, 1943–1948. - PubMed
-
- Chen, I.-H., Cheng, T., Wang, Y.-L., Huang, S.-J., Hsiao, Y.-H., Lai, Y.-T., Toh, S.-I., Chu, J., Rudolf, J. D. & Chang, C.-Y. (2022). ChemBioChem, 23, e202200563. - PubMed
-
- Dharavath, S., Raj, I. & Gourinath, S. (2017). Biochem. J. 474, 1221–1239. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

