Epigenetic silencing of OR and TAS2R genes expression in human orbitofrontal cortex at early stages of sporadic Alzheimer's disease
- PMID: 37405535
- PMCID: PMC10322771
- DOI: 10.1007/s00018-023-04845-1
Epigenetic silencing of OR and TAS2R genes expression in human orbitofrontal cortex at early stages of sporadic Alzheimer's disease
Abstract
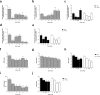
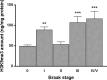
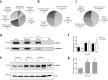
Modulation of brain olfactory (OR) and taste receptor (TASR) expression was recently reported in neurological diseases. However, there is still limited evidence of these genes' expression in the human brain and the transcriptional regulation mechanisms involved remain elusive. We explored the possible expression and regulation of selected OR and TASR in the human orbitofrontal cortex (OFC) of sporadic Alzheimer's disease (AD) and non-demented control specimens using quantitative real-time RT-PCR and ELISA. Global H3K9me3 amounts were measured on OFC total histone extracts, and H3K9me3 binding at each chemoreceptor locus was examined through native chromatin immunoprecipitation. To investigate the potential interactome of the repressive histone mark H3K9me3 in OFC specimens, native nuclear complex co-immunoprecipitation (Co-IP) was combined with reverse phase-liquid chromatography coupled to mass spectrometry analysis. Interaction between H3K9me3 and MeCP2 was validated by reciprocal Co-IP, and global MeCP2 levels were quantitated. We found that OR and TAS2R genes are expressed and markedly downregulated in OFC at early stages of sporadic AD, preceding the progressive reduction in their protein levels and the appearance of AD-associated neuropathology. The expression pattern did not follow disease progression suggesting transcriptional regulation through epigenetic mechanisms. We discovered an increase of OFC global H3K9me3 levels and a substantial enrichment of this repressive signature at ORs and TAS2Rs proximal promoter at early stages of AD, ultimately lost at advanced stages. We revealed the interaction between H3K9me3 and MeCP2 at early stages and found that MeCP2 protein is increased in sporadic AD. Findings suggest MeCP2 might be implicated in OR and TAS2R transcriptional regulation through interaction with H3K9me3, and as an early event, it may uncover a novel etiopathogenetic mechanism of sporadic AD.
Keywords: Alzheimer’s disease; Histone methylation; MeCP2; Olfactory receptors; Orbitofrontal cortex; Taste receptors.
© 2023. The Author(s).
Conflict of interest statement
The authors have no relevant financial or non-financial interests to disclose.
Figures