ZCCHC3 is a stress granule zinc knuckle protein that strongly suppresses LINE-1 retrotransposition
- PMID: 37405998
- PMCID: PMC10351740
- DOI: 10.1371/journal.pgen.1010795
ZCCHC3 is a stress granule zinc knuckle protein that strongly suppresses LINE-1 retrotransposition
Abstract
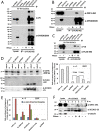
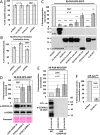
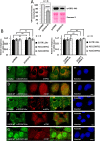
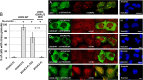
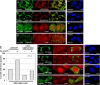
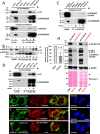
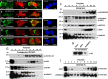
Retrotransposons have generated about half of the human genome and LINE-1s (L1s) are the only autonomously active retrotransposons. The cell has evolved an arsenal of defense mechanisms to protect against retrotransposition with factors we are only beginning to understand. In this study, we investigate Zinc Finger CCHC-Type Containing 3 (ZCCHC3), a gag-like zinc knuckle protein recently reported to function in the innate immune response to infecting viruses. We show that ZCCHC3 also severely restricts human retrotransposons and associates with the L1 ORF1p ribonucleoprotein particle. We identify ZCCHC3 as a bona fide stress granule protein, and its association with LINE-1 is further supported by colocalization with L1 ORF1 protein in stress granules, dense cytoplasmic aggregations of proteins and RNAs that contain stalled translation pre-initiation complexes and form when the cell is under stress. Our work also draws links between ZCCHC3 and the anti-viral and retrotransposon restriction factors Mov10 RISC Complex RNA Helicase (MOV10) and Zinc Finger CCCH-Type, Antiviral 1 (ZC3HAV1, also called ZAP). Furthermore, collective evidence from subcellular localization, co-immunoprecipitation, and velocity gradient centrifugation connects ZCCHC3 with the RNA exosome, a multi-subunit ribonuclease complex capable of degrading various species of RNA molecules and that has previously been linked with retrotransposon control.
Copyright: © 2023 Goodier et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

