Maternal Colonization Versus Nosocomial Transmission as the Source of Drug-Resistant Bloodstream Infection in an Indian Neonatal Intensive Care Unit: A Prospective Cohort Study
- PMID: 37406039
- PMCID: PMC10321698
- DOI: 10.1093/cid/ciad282
Maternal Colonization Versus Nosocomial Transmission as the Source of Drug-Resistant Bloodstream Infection in an Indian Neonatal Intensive Care Unit: A Prospective Cohort Study
Abstract
Background: Drug-resistant gram-negative (GN) pathogens are a common cause of neonatal sepsis in low- and middle-income countries. Identifying GN transmission patterns is vital to inform preventive efforts.
Methods: We conducted a prospective cohort study, 12 October 2018 to 31 October 2019 to describe the association of maternal and environmental GN colonization with bloodstream infection (BSI) among neonates admitted to a neonatal intensive care unit (NICU) in Western India. We assessed rectal and vaginal colonization in pregnant women presenting for delivery and colonization in neonates and the environment using culture-based methods. We also collected data on BSI for all NICU patients, including neonates born to unenrolled mothers. Organism identification, antibiotic susceptibility testing, and next-generation sequencing (NGS) were performed to compare BSI and related colonization isolates.
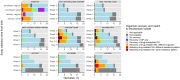
Results: Among 952 enrolled women who delivered, 257 neonates required NICU admission, and 24 (9.3%) developed BSI. Among mothers of neonates with GN BSI (n = 21), 10 (47.7%) had rectal, 5 (23.8%) had vaginal, and 10 (47.7%) had no colonization with resistant GN organisms. No maternal isolates matched the species and resistance pattern of associated neonatal BSI isolates. Thirty GN BSI were observed among neonates born to unenrolled mothers. Among 37 of 51 BSI with available NGS data, 21 (57%) showed a single nucleotide polymorphism distance of ≤5 to another BSI isolate.
Conclusions: Prospective assessment of maternal GN colonization did not demonstrate linkage to neonatal BSI. Organism-relatedness among neonates with BSI suggests nosocomial spread, highlighting the importance of NICU infection prevention and control practices to reduce GN BSI.
Keywords: antimicrobial resistance; newborn; sepsis.
© The Author(s) 2023. Published by Oxford University Press on behalf of Infectious Diseases Society of America.
Conflict of interest statement
Potential conflicts of interest. Y. C. M. reports payments to their institution from the CDC, consulting fees from Abbott STI Ad Board, honoraria payments from DKB Med on COVID, patents with TB polymer diagnostic, and holds a leadership role on the board with the Infectious Diseases Institution. They also report receipt of equipment from Hologic, Cepheid, Roche, ChemBio, Becton Dickinson, and miDiagnostics. A. G. reports funding from NIH, UNITAID, and CDC. They report participation in an advisory council with NIH/NIAID and the governing board with Indo US Science Technology and leadership roles with the IMPAACT Network TB Scientific Committee and the WHO MDR TB Guidelines Committee. J. J. reports funding from the CDC (grant number NU3HCK000001) and (grant number U54CK000617). They also report holding a position on the AAP Section on Neonatal-Pinatal Medicine Global Health Subcommittee and the Society for Healthcare Epidemiology of America Pediatric Leadership Council Steering Committee. M. R. Reports research and travel support from the CDC. V. M. reports funding support from NIH, NIAID, and CDC. A. M. reports research funding from CDC, NIH, AHRQ, and Merck. \ All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Figures


Similar articles
-
Gut Pathogen Colonization: A Risk Factor to Bloodstream Infections in Preterm Neonates Admitted in the Neonatal Intensive Care Unit - A Prospective Cohort Study.Neonatology. 2025;122(2):151-160. doi: 10.1159/000542335. Epub 2024 Dec 13. Neonatology. 2025. PMID: 39675351 Free PMC article.
-
High Burden of Bloodstream Infections Associated With Antimicrobial Resistance and Mortality in the Neonatal Intensive Care Unit in Pune, India.Clin Infect Dis. 2021 Jul 15;73(2):271-280. doi: 10.1093/cid/ciaa554. Clin Infect Dis. 2021. PMID: 32421763 Free PMC article.
-
Bloodstream infections and antimicrobial resistance patterns in a South African neonatal intensive care unit.Paediatr Int Child Health. 2014 May;34(2):108-14. doi: 10.1179/2046905513Y.0000000082. Epub 2013 Dec 6. Paediatr Int Child Health. 2014. PMID: 24621234
-
Neonatal bloodstream infections.Curr Opin Infect Dis. 2021 Oct 1;34(5):533-537. doi: 10.1097/QCO.0000000000000764. Curr Opin Infect Dis. 2021. PMID: 34261905 Review.
-
Colonization with antibiotic-resistant Gram-negative bacilli in the neonatal intensive care unit.Minerva Pediatr. 2003 Oct;55(5):385-93. Minerva Pediatr. 2003. PMID: 14608262 Review.
Cited by
-
The Pattern and Impact of Hospital-Acquired Infections and Its Outlook in India.Cureus. 2023 Nov 9;15(11):e48583. doi: 10.7759/cureus.48583. eCollection 2023 Nov. Cureus. 2023. PMID: 38090398 Free PMC article. Review.
-
Antibiotic Resistance: A Global Problem and the Need to Do More.Clin Infect Dis. 2023 Jul 5;77(Suppl 1):S1-S3. doi: 10.1093/cid/ciad226. Clin Infect Dis. 2023. PMID: 37406051 Free PMC article. No abstract available.
-
Strengthening hospital epidemiology & infection prevention research capacity in India to curb antimicrobial resistance.Indian J Med Res. 2024 Jan 1;159(1):7-9. doi: 10.4103/ijmr.ijmr_1919_23. Epub 2024 Mar 4. Indian J Med Res. 2024. PMID: 38344922 Free PMC article. No abstract available.
-
Risk Factors for Health Care-Associated Bloodstream Infections in NICUs.JAMA Netw Open. 2025 Mar 3;8(3):e251821. doi: 10.1001/jamanetworkopen.2025.1821. JAMA Netw Open. 2025. PMID: 40131271 Free PMC article.
-
Infectious Causes of Stillbirths: A Descriptive Etiological Study in Uganda.Open Forum Infect Dis. 2025 Mar 10;11(Suppl 3):S165-S172. doi: 10.1093/ofid/ofae606. eCollection 2024 Dec. Open Forum Infect Dis. 2025. PMID: 40070697 Free PMC article.
References
-
- Public Health Foundation of India . State of India's newborns. Available at: https://www.healthynewbornnetwork.org/hnn-content/uploads/SOIN_Report_20.... Accessed 10 August.
-
- Investigators of the Delhi Neonatal Infection Study (DeNIS) collaboration . Characterisation and antimicrobial resistance of sepsis pathogens in neonates born in tertiary care centres in Delhi, India: a cohort study. Lancet Glob Health 2016; 4:e752–60. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

