UHRF1 is a mediator of KRAS driven oncogenesis in lung adenocarcinoma
- PMID: 37407562
- PMCID: PMC10322837
- DOI: 10.1038/s41467-023-39591-2
UHRF1 is a mediator of KRAS driven oncogenesis in lung adenocarcinoma
Abstract
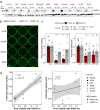
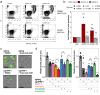
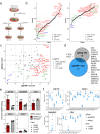
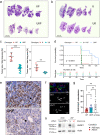
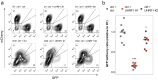
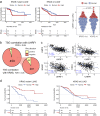
KRAS is a frequent driver in lung cancer. To identify KRAS-specific vulnerabilities in lung cancer, we performed RNAi screens in primary spheroids derived from a Kras mutant mouse lung cancer model and discovered an epigenetic regulator Ubiquitin-like containing PHD and RING finger domains 1 (UHRF1). In human lung cancer models UHRF1 knock-out selectively impaired growth and induced apoptosis only in KRAS mutant cells. Genome-wide methylation and gene expression analysis of UHRF1-depleted KRAS mutant cells revealed global DNA hypomethylation leading to upregulation of tumor suppressor genes (TSGs). A focused CRISPR/Cas9 screen validated several of these TSGs as mediators of UHRF1-driven tumorigenesis. In vivo, UHRF1 knock-out inhibited tumor growth of KRAS-driven mouse lung cancer models. Finally, in lung cancer patients high UHRF1 expression is anti-correlated with TSG expression and predicts worse outcomes for patients with KRAS mutant tumors. These results nominate UHRF1 as a KRAS-specific vulnerability and potential target for therapeutic intervention.
© 2023. The Author(s).
Conflict of interest statement
A.C. is a founder, equity holder, and consultant of DarwinHealth Inc., a company that has licensed some of the algorithms used in this manuscript from Columbia University. Columbia University is also an equity holder in DarwinHealth Inc. The remaining authors declare no competing interests.
Figures


References
-
- Liu J, Kang R, Tang D. The KRAS-G12C inhibitor: activity and resistance. Cancer Gene Ther. 2021;2021:1–4. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

