The cholesterol transporter NPC1 is essential for epigenetic regulation and maturation of oligodendrocyte lineage cells
- PMID: 37407594
- PMCID: PMC10322873
- DOI: 10.1038/s41467-023-39733-6
The cholesterol transporter NPC1 is essential for epigenetic regulation and maturation of oligodendrocyte lineage cells
Abstract
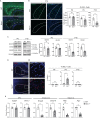
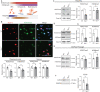
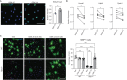
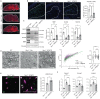
The intracellular cholesterol transporter NPC1 functions in late endosomes and lysosomes to efflux unesterified cholesterol, and its deficiency causes Niemann-Pick disease Type C, an autosomal recessive lysosomal disorder characterized by progressive neurodegeneration and early death. Here, we use single-nucleus RNA-seq on the forebrain of Npc1-/- mice at P16 to identify cell types and pathways affected early in pathogenesis. Our analysis uncovers significant transcriptional changes in the oligodendrocyte lineage during developmental myelination, accompanied by diminished maturation of myelinating oligodendrocytes. We identify upregulation of genes associated with neurogenesis and synapse formation in Npc1-/- oligodendrocyte lineage cells, reflecting diminished gene silencing by H3K27me3. Npc1-/- oligodendrocyte progenitor cells reproduce impaired maturation in vitro, and this phenotype is rescued by treatment with GSK-J4, a small molecule inhibitor of H3K27 demethylases. Moreover, mobilizing stored cholesterol in Npc1-/- mice by a single administration of 2-hydroxypropyl-β-cyclodextrin at P7 rescues myelination, epigenetic marks, and oligodendrocyte gene expression. Our findings highlight an important role for NPC1 in oligodendrocyte lineage maturation and epigenetic regulation, and identify potential targets for therapeutic intervention.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

