Multi-landmark alignment of genomic signals reveals conserved expression patterns across transcription start sites
- PMID: 37407625
- PMCID: PMC10322939
- DOI: 10.1038/s41598-023-37140-x
Multi-landmark alignment of genomic signals reveals conserved expression patterns across transcription start sites
Abstract
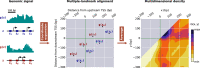
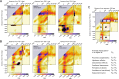
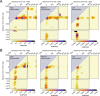
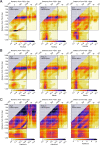
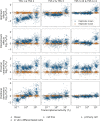
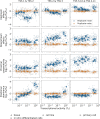
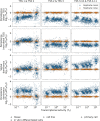
The prevalent one-dimensional alignment of genomic signals to a reference landmark is a cornerstone of current methods to study transcription and its DNA-dependent processes but it is prone to mask potential relations among multiple DNA elements. We developed a systematic approach to align genomic signals to multiple locations simultaneously by expanding the dimensionality of the genomic-coordinate space. We analyzed transcription in human and uncovered a complex dependence on the relative position of neighboring transcription start sites (TSSs) that is consistently conserved among cell types. The dependence ranges from enhancement to suppression of transcription depending on the relative distances to the TSSs, their intragenic position, and the transcriptional activity of the gene. Our results reveal a conserved hierarchy of alternative TSS usage within a previously unrecognized level of genomic organization and provide a general methodology to analyze complex functional relationships among multiple types of DNA elements.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures