A novel polypeptide CAPG-171aa encoded by circCAPG plays a critical role in triple-negative breast cancer
- PMID: 37408008
- PMCID: PMC10320902
- DOI: 10.1186/s12943-023-01806-x
A novel polypeptide CAPG-171aa encoded by circCAPG plays a critical role in triple-negative breast cancer
Abstract
Background: The treatment of Triple-negative breast cancer (TNBC) has always been challenging due to its heterogeneity and the absence of well-defined molecular targets. The present study aims to elucidate the role of protein-coding circRNAs in the etiology and carcinogenesis of TNBC.
Methods: CircRNA expression data in TNBC (GEO: GSE113230, GSE101123) were reanalyzed and then circCAPG was selected for further study. To identify the polypeptide-coding function of circCAPG, a series of experiments, such as Mass spectrometry and dual-luciferase reporter assays were conducted. Cell proliferation, apoptosis and metastasis parameters were determined to investigate the cancerous functions CAPG-171aa plays in both TNBC organoids and nude mice. Mechanistically, the relation between CAPG-171aa and STK38 in TNBC was verified by immunoprecipitation analyses and mass spectrometry. The interactions between SLU7 and its binding site on circCAPG were validated by RIP-qPCR experiments.
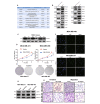
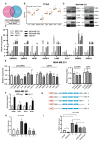
Results: In both TNBC clinical samples and cell lines, the expression level of circCAPG was identified to be higher compared with normal ones and positively correlated with the overall survival (n = 132) in a 10-year follow-up study, in which the area under the curve of receiver operating characteristic was 0.8723 with 100% specificity and 80% sensitivity. In addition, we found that circCAPG knockdown (KD) significantly inhibited the growth of TNBC organoids. Intriguingly, circCAPG can be translated into a polypeptide named CAPG-171aa which promotes tumor growh by disrupting the binding of serine/threonine kinase 38 (STK38) to SMAD-specific E3 ubiquitin protein ligase 1 (SMURF1) and thereby preventing MEKK2 ubiquitination and proteasomal degradation. Furthermore, we found that SLU7 Homolog- Splicing Factor (SLU7) can regulate the bio-generation of circCAPG through binding to the flanking Alu sequences of circRNA transcripts.
Conclusions: circCAPG significantly enhances the proliferation and metastasis of TNBC cells by encoding a novel polypeptide CAPG-171aa and afterwards activates MEKK2-MEK1/2-ERK1/2 pathway. Additionally, the formation of circCAPG is found to be mediated by SLU7. The present study provides innovative insight into the role of protein-coding circRNAs CAPG-171aa in TNBC, and its capacity to serve as a promising prognostic biomarker and potential therapeutic target in TNBC.
Keywords: CAPG-171aa; CircCAPG; MEKK2; SLU7; STK38; TNBC; Therapeutic target.
© 2023. The Author(s).
Conflict of interest statement
All authors declare that they have no competing interests.
Figures






References
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca-a Cancer Journal for Clinicians. 2021;71:209–249. - PubMed
-
- Liao CH, Zhang Y, Fan C, Herring LE, Liu J, Locasale JW, Takada M, Zhou J, Zurlo G, Hu LX, et al. Identification of BBOX1 as a therapeutic target in Triple-Negative breast Cancer. Cancer Discov. 2020;10:1706–21. doi: 10.1158/2159-8290.CD-20-0288. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

