p53 Regulates the Extent of Fibroblast Proliferation and Fibrosis in Left Ventricle Pressure Overload
- PMID: 37409456
- PMCID: PMC10361635
- DOI: 10.1161/CIRCRESAHA.121.320324
p53 Regulates the Extent of Fibroblast Proliferation and Fibrosis in Left Ventricle Pressure Overload
Abstract
Background: Cardiomyopathy is characterized by the pathological accumulation of resident cardiac fibroblasts that deposit ECM (extracellular matrix) and generate a fibrotic scar. However, the mechanisms that control the timing and extent of cardiac fibroblast proliferation and ECM production are not known, hampering the development of antifibrotic strategies to prevent heart failure.
Methods: We used the Tcf21 (transcription factor 21)MerCreMer mouse line for fibroblast-specific lineage tracing and p53 (tumor protein p53) gene deletion. We characterized cardiac physiology and used single-cell RNA-sequencing and in vitro studies to investigate the p53-dependent mechanisms regulating cardiac fibroblast cell cycle and fibrosis in left ventricular pressure overload induced by transaortic constriction.
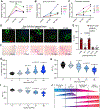
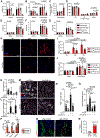
Results: Cardiac fibroblast proliferation occurs primarily between days 7 and 14 following transaortic constriction in mice, correlating with alterations in p53-dependent gene expression. p53 deletion in fibroblasts led to a striking accumulation of Tcf21-lineage cardiac fibroblasts within the normal proliferative window and precipitated a robust fibrotic response to left ventricular pressure overload. However, excessive interstitial and perivascular fibrosis does not develop until after cardiac fibroblasts exit the cell cycle. Single-cell RNA sequencing revealed p53 null fibroblasts unexpectedly express lower levels of genes encoding important ECM proteins while they exhibit an inappropriately proliferative phenotype. in vitro studies establish a role for p53 in suppressing the proliferative fibroblast phenotype, which facilitates the expression and secretion of ECM proteins. Importantly, Cdkn2a (cyclin-dependent kinase inhibitor 2a) expression and the p16Ink4a-retinoblastoma cell cycle control pathway is induced in p53 null cardiac fibroblasts, which may eventually contribute to cell cycle exit and fulminant scar formation.
Conclusions: This study reveals a mechanism regulating cardiac fibroblast accumulation and ECM secretion, orchestrated in part by p53-dependent cell cycle control that governs the timing and extent of fibrosis in left ventricular pressure overload.
Keywords: extracellular matrix; fibroblasts; fibrosis; heart; mice.
Conflict of interest statement
Figures






Comment in
-
p53 at the Intersection of Cardiac Fibroblast Proliferation and Activation: Answers and Questions.Circ Res. 2023 Jul 21;133(3):288-290. doi: 10.1161/CIRCRESAHA.123.323209. Epub 2023 Jul 20. Circ Res. 2023. PMID: 37471487 No abstract available.
References
-
- Russo I, Cavalera M, Huang S, Su Y, Hanna A, Chen B, Shinde AV, Conway SJ, Graff J and Frangogiannis NG. Protective Effects of Activated Myofibroblasts in the Pressure-Overloaded Myocardium Are Mediated Through Smad-Dependent Activation of a Matrix-Preserving Program. Circ Res. 2019;124:1214–1227. - PMC - PubMed
-
- Hill JA and Olson EN. Cardiac plasticity. N Engl J Med. 2008;358:1370–80. - PubMed
-
- Mozaffarian D, Benjamin EJ, Go AS, Arnett DK, Blaha MJ, Cushman M, Das SR, de Ferranti S, Despres JP, Fullerton HJ, et al. Heart Disease and Stroke Statistics-2016 Update: A Report From the American Heart Association. Circulation. 2016;133:e38–360. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

