DiscoVari: A Web-Based Precision Medicine Tool for Predicting Variant Pathogenicity in Cardiomyopathy- and Channelopathy-Associated Genes
- PMID: 37409478
- PMCID: PMC10527712
- DOI: 10.1161/CIRCGEN.122.003911
DiscoVari: A Web-Based Precision Medicine Tool for Predicting Variant Pathogenicity in Cardiomyopathy- and Channelopathy-Associated Genes
Abstract
Background: With genetic testing advancements, the burden of incidentally identified cardiac disease-associated gene variants is rising. These variants may carry a risk of sudden cardiac death, highlighting the need for accurate diagnostic interpretation. We sought to identify pathogenic hotspots in sudden cardiac death-associated genes using amino acid-level signal-to-noise (S:N) analysis and develop a web-based precision medicine tool, DiscoVari, to improve variant evaluation.
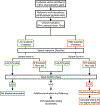
Methods: The minor allele frequency of putatively pathogenic variants was derived from cohort-based cardiomyopathy and channelopathy studies in the literature. We normalized disease-associated minor allele frequencies to rare variants in an ostensibly healthy population (Genome Aggregation Database) to calculate amino acid-level S:N. Amino acids with S:N above the gene-specific threshold were defined as hotspots. DiscoVari was built using JavaScript ES6 and using open-source JavaScript library ReactJS, web development framework Next.js, and JavaScript runtime NodeJS. We validated the ability of DiscoVari to identify pathogenic variants using variants from ClinVar and individuals clinically evaluated at the Duke University Hospitals with cardiac genetic testing.
Results: We developed DiscoVari as an internet-based tool for S:N-based variant hotspots. Upon validation, a higher proportion of ClinVar likely pathogenic/pathogenic variants localized to DiscoVari hotspots (43.1%) than likely benign/benign variants (17.8%; P<0.0001). Further, 75.3% of ClinVar variants reclassified to likely pathogenic/pathogenic were in hotspots, compared with 41.3% of those reclassified as variants of uncertain significance (P<0.0001) and 23.4% of those reclassified as likely benign/benign (P<0.0001). Of the clinical cohort variants, 73.1% of likely pathogenic/pathogenic were in hotspots, compared with 0.0% of likely benign/benign (P<0.01).
Conclusions: DiscoVari reliably identifies disease-susceptible amino acid residues to evaluate variants by searching amino acid-specific S:N ratios.
Keywords: channelopathy; exome sequencing; long QT syndrome; probability; syncope.
Conflict of interest statement
Figures




References
-
- Kurzlechner LM, Jones EG, Berkman AM, Tadros HJ, Rosenfeld JA, Yang Y, Tunuguntla H, Allen HD, Kim JJ, Landstrom AP. Signal-to-Noise Analysis Can Inform the Likelihood That Incidentally Identified Variants in Sarcomeric Genes Are Associated with Pediatric Cardiomyopathy. J Pers Med 2022;12. doi: 10.3390/jpm12050733 - DOI - PMC - PubMed
-
- Miller DT, Lee K, Chung WK, Gordon AS, Herman GE, Klein TE, Stewart DR, Amendola LM, Adelman K, Bale SJ, et al. ACMG SF v3.0 list for reporting of secondary findings in clinical exome and genome sequencing: a policy statement of the American College of Medical Genetics and Genomics (ACMG). Genet Med 2021;23:1381–1390. doi: 10.1038/s41436-021-01172-3 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

