S2P intramembrane protease RseP degrades small membrane proteins and suppresses the cytotoxicity of intrinsic toxin HokB
- PMID: 37409810
- PMCID: PMC10470546
- DOI: 10.1128/mbio.01086-23
S2P intramembrane protease RseP degrades small membrane proteins and suppresses the cytotoxicity of intrinsic toxin HokB
Abstract
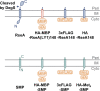
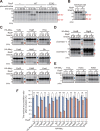
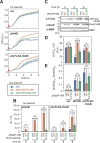
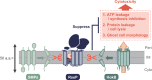
The site2-protease (S2P) family of intramembrane proteases (IMPs) is conserved in all kingdoms of life and cleaves transmembrane proteins within the membrane to regulate and maintain various cellular activities. RseP, an Escherichia coli S2P peptidase, is involved in the regulation of gene expression through the regulated cleavage of the two target membrane proteins (RseA and FecR) and in membrane quality control through the proteolytic elimination of remnant signal peptides. RseP is expected to have additional substrates and to be involved in other cellular processes. Recent studies have shown that cells express small membrane proteins (SMPs; single-spanning membrane proteins of approximately 50-100 amino acid residues) with crucial cellular functions. However, little is known about their metabolism, which affects their functions. This study investigated the possible RseP-catalyzed cleavage of E. coli SMPs based on the apparent similarity of the sizes and structures of SMPs to those of remnant signal peptides. We screened SMPs cleaved by RseP in vivo and in vitro and identified 14 SMPs, including HokB, an endogenous toxin that induces persister formation, as potential substrates. We demonstrated that RseP suppresses the cytotoxicity and biological functions of HokB. The identification of several SMPs as novel potential substrates of RseP provides a clue to a comprehensive understanding of the cellular roles of RseP and other S2P peptidases and highlights a novel aspect of the regulation of SMPs. IMPORTANCE Membrane proteins play an important role in cell activity and survival. Thus, understanding their dynamics, including proteolytic degradation, is crucial. E. coli RseP, an S2P family intramembrane protease, cleaves membrane proteins to regulate gene expression in response to environmental changes and to maintain membrane quality. To identify novel substrates of RseP, we screened small membrane proteins (SMPs), a group of proteins that have recently been shown to have diverse cellular functions, and identified 14 potential substrates. We also showed that RseP suppresses the cytotoxicity of the intrinsic toxin, HokB, an SMP that has been reported to induce persister cell formation, by degrading it. These findings provide new insights into the cellular roles of S2P peptidases and the functional regulation of SMPs.
Keywords: extracytoplasmic stress response; membrane protease; proteostasis; regulated intramembrane proteolysis; zinc metallopeptidase.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
The Escherichia coli S2P intramembrane protease RseP regulates ferric citrate uptake by cleaving the sigma factor regulator FecR.J Biol Chem. 2021 Jan-Jun;296:100673. doi: 10.1016/j.jbc.2021.100673. Epub 2021 Apr 16. J Biol Chem. 2021. PMID: 33865858 Free PMC article.
-
Involvement of a Membrane-Bound Amphiphilic Helix in Substrate Discrimination and Binding by an Escherichia coli S2P Peptidase RseP.Front Microbiol. 2020 Nov 27;11:607381. doi: 10.3389/fmicb.2020.607381. eCollection 2020. Front Microbiol. 2020. PMID: 33329500 Free PMC article.
-
Substrate recognition and binding by RseP, an Escherichia coli intramembrane protease.J Biol Chem. 2008 Apr 11;283(15):9562-70. doi: 10.1074/jbc.M709984200. Epub 2008 Feb 11. J Biol Chem. 2008. PMID: 18268014
-
Biochemical Characterization of Function and Structure of RseP, an Escherichia coli S2P Protease.Methods Enzymol. 2017;584:1-33. doi: 10.1016/bs.mie.2016.09.044. Epub 2016 Oct 31. Methods Enzymol. 2017. PMID: 28065260 Review.
-
New insights into S2P signaling cascades: regulation, variation, and conservation.Protein Sci. 2010 Nov;19(11):2015-30. doi: 10.1002/pro.496. Protein Sci. 2010. PMID: 20836086 Free PMC article. Review.
Cited by
-
Toxin-antitoxin genes are differentially expressed in Escherichia coli relA and spoT mutans cultured under nitrogen, fatty acid, or carbon starvation conditions.Front Microbiol. 2025 Jan 17;15:1528825. doi: 10.3389/fmicb.2024.1528825. eCollection 2024. Front Microbiol. 2025. PMID: 39895937 Free PMC article.
-
Cryo-EM structure of the bacterial intramembrane metalloprotease RseP in the substrate-bound state.Sci Adv. 2025 Feb 28;11(9):eadu0925. doi: 10.1126/sciadv.adu0925. Epub 2025 Feb 26. Sci Adv. 2025. PMID: 40009668 Free PMC article.
-
Cleavage cascade of the sigma regulator FecR orchestrates TonB-dependent signal transduction.Proc Natl Acad Sci U S A. 2025 Apr 22;122(16):e2500366122. doi: 10.1073/pnas.2500366122. Epub 2025 Apr 17. Proc Natl Acad Sci U S A. 2025. PMID: 40244679
-
Site-2 protease Sll0528 interacts with RbcR to regulate carbon/nitrogen homeostasis in the cyanobacterium Synechocystis sp. PCC 6803.Front Microbiol. 2025 Apr 9;16:1556583. doi: 10.3389/fmicb.2025.1556583. eCollection 2025. Front Microbiol. 2025. PMID: 40270807 Free PMC article.
-
Maintaining the Integral Membrane Proteome: Revisiting the Functional Repertoire of Integral Membrane Proteases.Chembiochem. 2025 May 5;26(9):e202500048. doi: 10.1002/cbic.202500048. Epub 2025 Mar 18. Chembiochem. 2025. PMID: 40056010 Free PMC article. Review.
References
Grants and funding
- JP21J15841/MEXT | Japan Society for the Promotion of Science (JSPS)
- JP26291016/MEXT | Japan Society for the Promotion of Science (JSPS)
- JP19H03170/MEXT | Japan Society for the Promotion of Science (JSPS)
- JP22H02561/MEXT | Japan Society for the Promotion of Science (JSPS)
- JP16am0101020/Platform Project for Supporting in Drug Discovery and Life Science Research form the Japan Agency for Medical Research and Development
- N.A./Cooperative Research Program (Joint Usage/Research Centerprogram) of the Institute for Frontier Life and Medical Sciences Kyoto University
- JP19K06562/MEXT | Japan Society for the Promotion of Science (JSPS)
- JP22K06142/MEXT | Japan Society for the Promotion of Science (JSPS)
- N.A./Institute for Frontier Life and Medical Sciences, Kyoto University for INFRONT Office of Director's Research Grant Program (2020)
- N.A./Institute for Frontier Life and Medical Sciences, Kyoto University for INFRONT Office of Director's Research Grant Program (2021)
- G-2022-2-108/Institute for Fermentation, Osaka (IFO)
- JP18H02404/MEXT | Japan Society for the Promotion of Science (JSPS)
- JP22H02571/MEXT | Japan Society for the Promotion of Science (JSPS)
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

