Variations in Genes Encoding Human Papillomavirus Binding Receptors and Susceptibility to Cervical Precancer
- PMID: 37410084
- PMCID: PMC10472094
- DOI: 10.1158/1055-9965.EPI-23-0300
Variations in Genes Encoding Human Papillomavirus Binding Receptors and Susceptibility to Cervical Precancer
Abstract
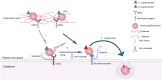
Background: Cervical cancer oncogenesis starts with human papillomavirus (HPV) cell entry after binding to host cell surface receptors; however, the mechanism is not fully known. We examined polymorphisms in receptor genes hypothesized to be necessary for HPV cell entry and assessed their associations with clinical progression to precancer.
Methods: African American women (N = 1,728) from the MACS/WIHS Combined Cohort Study were included. Two case-control study designs were used-cases with histology-based precancer (CIN3+) and controls without; and cases with cytology-based precancer [high-grade squamous intraepithelial lesions (HSIL)] and controls without. SNPs in candidate genes (SDC1, SDC2, SDC3, SDC4, GPC1, GPC2, GPC3, GPC4, GPC5, GPC6, and ITGA6) were genotyped using an Illumina Omni2.5-quad beadchip. Logistic regression was used to assess the associations in all participants and by HPV genotypes, after adjusting for age, human immunodeficiency virus serostatus, CD4 T cells, and three principal components for ancestry.
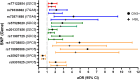
Results: Minor alleles in SNPs rs77122854 (SDC3), rs73971695, rs79336862 (ITGA6), rs57528020, rs201337456, rs11987725 (SDC2), rs115880588, rs115738853, and rs9301825 (GPC5) were associated with increased odds of both CIN3+ and HSIL, whereas, rs35927186 (GPC5) was found to decrease the odds for both outcomes (P value ≤ 0.01). Among those infected with Alpha-9 HPV types, rs722377 (SDC3), rs16860468, rs2356798 (ITGA6), rs11987725 (SDC2), and rs3848051 (GPC5) were associated with increased odds of both precancer outcomes.
Conclusions: Polymorphisms in genes that encode binding receptors for HPV cell entry may play a role in cervical precancer progression.
Impact: Our findings are hypothesis generating and support further exploration of mechanisms of HPV entry genes that may help prevent progression to cervical precancer.
©2023 The Authors; Published by the American Association for Cancer Research.
Figures
References
-
- zur Hausen H. Papillomaviruses in the causation of human cancers - a brief historical account. Virology 2009;384:260–5. - PubMed
-
- Walboomers JM, Jacobs MV, Manos MM, Bosch FX, Kummer JA, Shah KV, et al. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol 1999;189:12–19. - PubMed
-
- Ho GY, Bierman R, Beardsley L, Chang CJ, Burk RD. Natural history of cervicovaginal papillomavirus infection in young women. N Engl J Med 1998;338:423–8. - PubMed
-
- Moscicki AB, Shiboski S, Broering J, Powell K, Clayton L, Jay N, et al. The natural history of human papillomavirus infection as measured by repeated DNA testing in adolescent and young women. J Pediatr 1998;132:277–84. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U01 HL146245/HL/NHLBI NIH HHS/United States
- U01 HL146208/HL/NHLBI NIH HHS/United States
- UL1 TR001409/TR/NCATS NIH HHS/United States
- KL2 TR001432/TR/NCATS NIH HHS/United States
- U01 HL146192/HL/NHLBI NIH HHS/United States
- U01 HL146242/HL/NHLBI NIH HHS/United States
- TL1 TR001431/TR/NCATS NIH HHS/United States
- U01 HL146193/HL/NHLBI NIH HHS/United States
- U01 AI103401/AI/NIAID NIH HHS/United States
- U54 CA254568/CA/NCI NIH HHS/United States
- U01 HL146194/HL/NHLBI NIH HHS/United States
- U01 HL146241/HL/NHLBI NIH HHS/United States
- P30 AI027767/AI/NIAID NIH HHS/United States
- P30 AI050409/AI/NIAID NIH HHS/United States
- U01 HL146333/HL/NHLBI NIH HHS/United States
- U01 HL146205/HL/NHLBI NIH HHS/United States
- P30 MH116867/MH/NIMH NIH HHS/United States
- P30 AI073961/AI/NIAID NIH HHS/United States
- U01 HL146201/HL/NHLBI NIH HHS/United States
- U01 HL146204/HL/NHLBI NIH HHS/United States
- U01 HL146202/HL/NHLBI NIH HHS/United States
- UL1 TR001881/TR/NCATS NIH HHS/United States
- UL1 TR000004/TR/NCATS NIH HHS/United States
- U01 HL146240/HL/NHLBI NIH HHS/United States
- U01 HL146203/HL/NHLBI NIH HHS/United States
- UL1 TR003098/TR/NCATS NIH HHS/United States
- P30 AI050410/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

