Rapid, One-Step Sample Processing for Label-Free Single-Cell Proteomics
- PMID: 37410391
- PMCID: PMC11017373
- DOI: 10.1021/jasms.3c00159
Rapid, One-Step Sample Processing for Label-Free Single-Cell Proteomics
Abstract
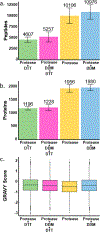
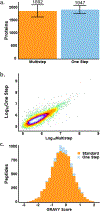
Sample preparation for single-cell proteomics is generally performed in a one-pot workflow with multiple dispensing and incubation steps. These hours-long processes can be labor intensive and lead to long sample-to-answer times. Here we report a sample preparation method that achieves cell lysis, protein denaturation, and digestion in 1 h using commercially available high-temperature-stabilized proteases with a single reagent dispensing step. Four different one-step reagent compositions were evaluated, and the mixture providing the highest proteome coverage was compared to the previously employed multistep workflow. The one-step preparation increases proteome coverage relative to the previous multistep workflow while minimizing labor input and the possibility of human error. We also compared sample recovery between previously used microfabricated glass nanowell chips and injection-molded polypropylene chips and found the polypropylene provided improved proteome coverage. Combined, the one-step sample preparation and the polypropylene substrates enabled the identification of an average of nearly 2400 proteins per cell using a standard data-dependent workflow with Orbitrap mass spectrometers. These advances greatly simplify sample preparation for single-cell proteomics and broaden accessibility with no compromise in terms of proteome coverage.
Conflict of interest statement
The authors declare the following competing financial interest(s): R.T.K. has a financial interest in MicrOmics Technologies LLC, which provided nanoelectrospray emitters used in this study.
Figures


References
-
- Stuart T; Satija R Integrative single-cell analysis. Nat. Rev. Genet. 2019, 20 (5), 257–272. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

