α7nAChR activation in AT2 cells promotes alveolar regeneration through WNT7B signaling in acute lung injury
- PMID: 37410546
- PMCID: PMC10445688
- DOI: 10.1172/jci.insight.162547
α7nAChR activation in AT2 cells promotes alveolar regeneration through WNT7B signaling in acute lung injury
Abstract
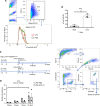
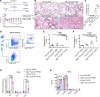
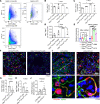
Reducing inflammatory damage and improving alveolar epithelium regeneration are two key approaches to promoting lung repair in acute lung injury/acute respiratory distress syndrome (ALI/ARDS). Stimulation of cholinergic α7 nicotinic acetylcholine receptor (α7nAChR, coded by Chrna7) signaling could dampen lung inflammatory injury. However, whether activation of α7nAChR in alveolar type II (AT2) cells promotes alveolar epithelial injury repair and underlying mechanisms is elusive. Here, we found that α7nAChR was expressed on AT2 cells and was upregulated in response to LPS-induced ALI. Meanwhile, deletion of Chrna7 in AT2 cells impeded lung repair process and worsened lung inflammation in ALI. Using in vivo AT2 lineage-labeled mice and ex vivo AT2 cell-derived alveolar organoids, we demonstrated that activation of α7nAChR expressed on AT2 cells improved alveolar regeneration by promoting AT2 cells to proliferate and subsequently differentiate toward alveolar type I cells. Then, we screened out the WNT7B signaling pathway by the RNA-Seq analysis of in vivo AT2 lineage-labeled cells and further confirmed its indispensability for α7nAChR activation-mediated alveolar epithelial proliferation and differentiation. Thus, we have identified a potentially unrecognized pathway in which cholinergic α7nAChR signaling determines alveolar regeneration and repair, which might provide us a novel therapeutic target for combating ALI.
Keywords: Adult stem cells; Pulmonology; Respiration; Stem cells.
Figures




Similar articles
-
VNS-mediated α7nAChR signaling promotes SPM synthesis via regulation of netrin-1 expression during LPS-induced ALI.FASEB J. 2024 Jan;38(1):e9664. doi: 10.1096/fj.202301623R. FASEB J. 2024. PMID: 38038805
-
Electroacupuncture Alleviates LPS-Induced ARDS Through α7 Nicotinic Acetylcholine Receptor-Mediated Inhibition of Ferroptosis.Front Immunol. 2022 Feb 10;13:832432. doi: 10.3389/fimmu.2022.832432. eCollection 2022. Front Immunol. 2022. PMID: 35222419 Free PMC article.
-
Vagal-α7nAChR signaling promotes lung stem cells regeneration via fibroblast growth factor 10 during lung injury repair.Stem Cell Res Ther. 2020 Jun 10;11(1):230. doi: 10.1186/s13287-020-01757-w. Stem Cell Res Ther. 2020. PMID: 32522255 Free PMC article.
-
Dual effects of supplemental oxygen on pulmonary infection, inflammatory lung injury, and neuromodulation in aging and COVID-19.Free Radic Biol Med. 2022 Sep;190:247-263. doi: 10.1016/j.freeradbiomed.2022.08.004. Epub 2022 Aug 11. Free Radic Biol Med. 2022. PMID: 35964839 Free PMC article. Review.
-
Interplay between pulmonary epithelial stem cells and innate immune cells contribute to the repair and regeneration of ALI/ARDS.Transl Res. 2024 Oct;272:111-125. doi: 10.1016/j.trsl.2024.05.012. Epub 2024 Jun 17. Transl Res. 2024. PMID: 38897427 Review.
Cited by
-
Targeting α7 Nicotinic Acetylcholine Receptor for Modulating the Neuroinflammation of Dry Eye Disease Via Macrophages.Invest Ophthalmol Vis Sci. 2025 May 1;66(5):13. doi: 10.1167/iovs.66.5.13. Invest Ophthalmol Vis Sci. 2025. PMID: 40327011 Free PMC article.
-
Neuroimmune recognition and regulation in the respiratory system.Eur Respir Rev. 2024 Jun 26;33(172):240008. doi: 10.1183/16000617.0008-2024. Print 2024 Apr. Eur Respir Rev. 2024. PMID: 38925790 Free PMC article. Review.
-
Exploring the mechanism of cytisine in treating respiratory depression following venomous snake bites based on network pharmacology and molecular docking.Am J Transl Res. 2024 Apr 15;16(4):1442-1453. doi: 10.62347/MCPD8618. eCollection 2024. Am J Transl Res. 2024. PMID: 38715819 Free PMC article.
-
Characterization of dipyridamole as a novel ferroptosis inhibitor and its therapeutic potential in acute respiratory distress syndrome management.Theranostics. 2024 Oct 21;14(18):6947-6968. doi: 10.7150/thno.102318. eCollection 2024. Theranostics. 2024. PMID: 39629132 Free PMC article.
-
Activation of nicotinic acetylcholine receptor α7 subunit limits Zika viral infection via promoting autophagy and ferroptosis.Mol Ther. 2024 Aug 7;32(8):2641-2661. doi: 10.1016/j.ymthe.2024.05.037. Epub 2024 May 31. Mol Ther. 2024. PMID: 38822526 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

