A systematic CRISPR screen reveals redundant and specific roles for Dscam1 isoform diversity in neuronal wiring
- PMID: 37410725
- PMCID: PMC10325099
- DOI: 10.1371/journal.pbio.3002197
A systematic CRISPR screen reveals redundant and specific roles for Dscam1 isoform diversity in neuronal wiring
Abstract
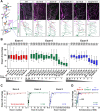
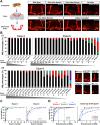
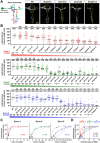
Drosophila melanogaster Down syndrome cell adhesion molecule 1 (Dscam1) encodes 19,008 diverse ectodomain isoforms via the alternative splicing of exon 4, 6, and 9 clusters. However, whether individual isoforms or exon clusters have specific significance is unclear. Here, using phenotype-diversity correlation analysis, we reveal the redundant and specific roles of Dscam1 diversity in neuronal wiring. A series of deletion mutations were performed from the endogenous locus harboring exon 4, 6, or 9 clusters, reducing to 396 to 18,612 potential ectodomain isoforms. Of the 3 types of neurons assessed, dendrite self/non-self discrimination required a minimum number of isoforms (approximately 2,000), independent of exon clusters or isoforms. In contrast, normal axon patterning in the mushroom body and mechanosensory neurons requires many more isoforms that tend to associate with specific exon clusters or isoforms. We conclude that the role of the Dscam1 diversity in dendrite self/non-self discrimination is nonspecifically mediated by its isoform diversity. In contrast, a separate role requires variable domain- or isoform-related functions and is essential for other neurodevelopmental contexts, such as axonal growth and branching. Our findings shed new light on a general principle for the role of Dscam1 diversity in neuronal wiring.
Copyright: © 2023 Dong et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




Similar articles
-
Self-avoidance alone does not explain the function of Dscam1 in mushroom body axonal wiring.Curr Biol. 2022 Jul 11;32(13):2908-2920.e4. doi: 10.1016/j.cub.2022.05.030. Epub 2022 Jun 2. Curr Biol. 2022. PMID: 35659864
-
Intron-targeted mutagenesis reveals roles for Dscam1 RNA pairing architecture-driven splicing bias in neuronal wiring.Cell Rep. 2021 Jul 13;36(2):109373. doi: 10.1016/j.celrep.2021.109373. Cell Rep. 2021. PMID: 34260933
-
Robust discrimination between self and non-self neurites requires thousands of Dscam1 isoforms.Nature. 2009 Oct 1;461(7264):644-8. doi: 10.1038/nature08431. Nature. 2009. PMID: 19794492 Free PMC article.
-
Revisiting Dscam diversity: lessons from clustered protocadherins.Cell Mol Life Sci. 2019 Feb;76(4):667-680. doi: 10.1007/s00018-018-2951-4. Epub 2018 Oct 20. Cell Mol Life Sci. 2019. PMID: 30343321 Free PMC article. Review.
-
Dscam-mediated cell recognition regulates neural circuit formation.Annu Rev Cell Dev Biol. 2008;24:597-620. doi: 10.1146/annurev.cellbio.24.110707.175250. Annu Rev Cell Dev Biol. 2008. PMID: 18837673 Free PMC article. Review.
Cited by
-
Functional insights into immunoglobulin superfamily proteins in invertebrate neurobiology and immunity.Front Immunol. 2025 Apr 2;16:1552151. doi: 10.3389/fimmu.2025.1552151. eCollection 2025. Front Immunol. 2025. PMID: 40242768 Free PMC article. Review.
-
Exploring perspectives of Dscam for cognitive deficits: a review of multifunction for regulating neural wiring in homeostasis.Front Mol Neurosci. 2025 May 2;18:1575348. doi: 10.3389/fnmol.2025.1575348. eCollection 2025. Front Mol Neurosci. 2025. PMID: 40385132 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

