The ubiquitin-protein ligase MIEL1 localizes to peroxisomes to promote seedling oleosin degradation and lipid droplet mobilization
- PMID: 37410814
- PMCID: PMC10629534
- DOI: 10.1073/pnas.2304870120
The ubiquitin-protein ligase MIEL1 localizes to peroxisomes to promote seedling oleosin degradation and lipid droplet mobilization
Abstract
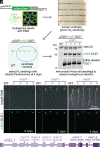
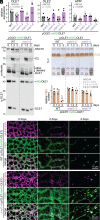
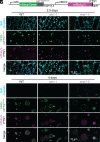
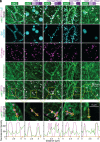
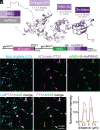
Lipid droplets are organelles conserved across eukaryotes that store and release neutral lipids to regulate energy homeostasis. In oilseed plants, fats stored in seed lipid droplets provide fixed carbon for seedling growth before photosynthesis begins. As fatty acids released from lipid droplet triacylglycerol are catabolized in peroxisomes, lipid droplet coat proteins are ubiquitinated, extracted, and degraded. In Arabidopsis seeds, the predominant lipid droplet coat protein is OLEOSIN1 (OLE1). To identify genes modulating lipid droplet dynamics, we mutagenized a line expressing mNeonGreen-tagged OLE1 expressed from the OLE1 promoter and isolated mutants with delayed oleosin degradation. From this screen, we identified four miel1 mutant alleles. MIEL1 (MYB30-interacting E3 ligase 1) targets specific MYB transcription factors for degradation during hormone and pathogen responses [D. Marino et al., Nat. Commun. 4, 1476 (2013); H. G. Lee and P. J. Seo, Nat. Commun. 7, 12525 (2016)] but had not been implicated in lipid droplet dynamics. OLE1 transcript levels were unchanged in miel1 mutants, indicating that MIEL1 modulates oleosin levels posttranscriptionally. When overexpressed, fluorescently tagged MIEL1 reduced oleosin levels, causing very large lipid droplets. Unexpectedly, fluorescently tagged MIEL1 localized to peroxisomes. Our data suggest that MIEL1 ubiquitinates peroxisome-proximal seed oleosins, targeting them for degradation during seedling lipid mobilization. The human MIEL1 homolog (PIRH2; p53-induced protein with a RING-H2 domain) targets p53 and other proteins for degradation and promotes tumorigenesis [A. Daks et al., Cells 11, 1515 (2022)]. When expressed in Arabidopsis, human PIRH2 also localized to peroxisomes, hinting at a previously unexplored role for PIRH2 in lipid catabolism and peroxisome biology in mammals.
Keywords: Arabidopsis thaliana; E3 ubiquitin ligase; PIRH2; lipid droplet; peroxisome.
Conflict of interest statement
The authors declare no competing interest.
Figures




Similar articles
-
Ubiquitin-Mediated Proteasomal Degradation of Oleosins is Involved in Oil Body Mobilization During Post-Germinative Seedling Growth in Arabidopsis.Plant Cell Physiol. 2015 Jul;56(7):1374-87. doi: 10.1093/pcp/pcv056. Epub 2015 Apr 22. Plant Cell Physiol. 2015. PMID: 25907570
-
The Arabidopsis MIEL1 E3 ligase negatively regulates ABA signalling by promoting protein turnover of MYB96.Nat Commun. 2016 Sep 12;7:12525. doi: 10.1038/ncomms12525. Nat Commun. 2016. PMID: 27615387 Free PMC article.
-
The MIEL1 E3 Ubiquitin Ligase Negatively Regulates Cuticular Wax Biosynthesis in Arabidopsis Stems.Plant Cell Physiol. 2017 Jul 1;58(7):1249-1259. doi: 10.1093/pcp/pcx065. Plant Cell Physiol. 2017. PMID: 28838126
-
Exploiting lipid droplet metabolic pathway to foster lipid production: oleosin in focus.Plant Cell Rep. 2024 Dec 26;44(1):12. doi: 10.1007/s00299-024-03390-w. Plant Cell Rep. 2024. PMID: 39724216 Review.
-
Lipid droplet mobilization: The different ways to loosen the purse strings.Biochimie. 2016 Jan;120:17-27. doi: 10.1016/j.biochi.2015.07.010. Epub 2015 Jul 15. Biochimie. 2016. PMID: 26187474 Review.
Cited by
-
Degeneration of oil bodies by rough endoplasmic reticulum -associated protein during seed germination in Cannabis sativa.AoB Plants. 2023 Nov 22;15(6):plad082. doi: 10.1093/aobpla/plad082. eCollection 2023 Dec. AoB Plants. 2023. PMID: 38094511 Free PMC article.
-
The Atypical Pectin Methylesterase Family Member PME31 Promotes Seedling Lipid Droplet Utilization.Plant Direct. 2025 Apr 9;9(4):e70054. doi: 10.1002/pld3.70054. eCollection 2025 Apr. Plant Direct. 2025. PMID: 40212536 Free PMC article.
-
Identifying and characterizing a missing peroxin-PEX8-in Arabidopsis thaliana.Plant Cell. 2025 Jul 1;37(7):koaf166. doi: 10.1093/plcell/koaf166. Plant Cell. 2025. PMID: 40577590 Free PMC article.
-
Global impacts of peroxisome and pexophagy dysfunction revealed through multi-omics analyses of lon2 and atg2 mutants.Plant J. 2024 Dec;120(6):2563-2583. doi: 10.1111/tpj.17129. Epub 2024 Nov 11. Plant J. 2024. PMID: 39526456 Free PMC article.
-
Profile of Bonnie Bartel.Proc Natl Acad Sci U S A. 2023 Oct 10;120(41):e2314758120. doi: 10.1073/pnas.2314758120. Epub 2023 Oct 2. Proc Natl Acad Sci U S A. 2023. PMID: 37782802 Free PMC article. No abstract available.
References
-
- Lundquist P. K., Shivaiah K.-K., Espinoza-Corral R., Lipid droplets throughout the evolutionary tree. Prog. Lipid Res. 78, 101029 (2020). - PubMed
-
- Guzha A., Whitehead P., Ischebeck T., Chapman K. D., Lipid droplets: packing hydrophobic molecules within the aqueous cytoplasm. Annu. Rev. Plant Biol. 74, 195–223 (2023). - PubMed
-
- Ischebeck T., Krawczyk H. E., Mullen R. T., Dyer J. M., Chapman K. D., Lipid droplets in plants and algae: Distribution, formation, turnover and function. Semin. Cell Dev. Biol. 108, 82–93 (2020). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

