VariantHunter: a method and tool for fast detection of emerging SARS-CoV-2 variants
- PMID: 37410916
- PMCID: PMC10325486
- DOI: 10.1093/database/baad044
VariantHunter: a method and tool for fast detection of emerging SARS-CoV-2 variants
Abstract
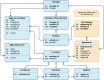
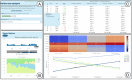
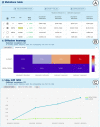
With the progression of the COVID-19 pandemic, large datasets of SARS-CoV-2 genome sequences were collected to closely monitor the evolution of the virus and identify the novel variants/strains. By analyzing genome sequencing data, health authorities can 'hunt' novel emerging variants of SARS-CoV-2 as early as possible, and then monitor their evolution and spread. We designed VariantHunter, a highly flexible and user-friendly tool for systematically monitoring the evolution of SARS-CoV-2 at global and regional levels. In VariantHunter, amino acid changes are analyzed over an interval of 4 weeks in an arbitrary geographical area (continent, country, or region); for every week in the interval, the prevalence is computed and changes are ranked based on their increase or decrease in prevalence. VariantHunter supports two main types of analysis: lineage-independent and lineage-specific. The former considers all the available data and aims to discover new viral variants. The latter evaluates specific lineages/viral variants to identify novel candidate designations (sub-lineages and sub-variants). Both analyses use simple statistics and visual representations (diffusion charts and heatmaps) to track viral evolution. A dataset explorer allows users to visualize available data and refine their selection. VariantHunter is a web application free to all users. The two types of supported analysis (lineage-independent and lineage-specific) allow user-friendly monitoring of the viral evolution, empowering genomic surveillance without requiring any computational background. Database URL http://gmql.eu/variant_hunter/.
© The Author(s) 2023. Published by Oxford University Press.
Conflict of interest statement
None declared.
Figures

Similar articles
-
ViruClust: direct comparison of SARS-CoV-2 genomes and genetic variants in space and time.Bioinformatics. 2022 Mar 28;38(7):1988-1994. doi: 10.1093/bioinformatics/btac030. Bioinformatics. 2022. PMID: 35040923
-
Wastewater Genomic Surveillance Captures Early Detection of Omicron in Utah.Microbiol Spectr. 2023 Jun 15;11(3):e0039123. doi: 10.1128/spectrum.00391-23. Epub 2023 May 8. Microbiol Spectr. 2023. PMID: 37154725 Free PMC article.
-
Early detection of SARS-CoV-2 variants through dynamic co-mutation network surveillance.Front Public Health. 2023 Jan 23;11:1015969. doi: 10.3389/fpubh.2023.1015969. eCollection 2023. Front Public Health. 2023. PMID: 36755900 Free PMC article.
-
One year into the pandemic: Short-term evolution of SARS-CoV-2 and emergence of new lineages.Infect Genet Evol. 2021 Aug;92:104869. doi: 10.1016/j.meegid.2021.104869. Epub 2021 Apr 26. Infect Genet Evol. 2021. PMID: 33915216 Free PMC article. Review.
-
Molecular evolution of SARS-CoV-2 from December 2019 to August 2022.J Med Virol. 2023 Jan;95(1):e28366. doi: 10.1002/jmv.28366. J Med Virol. 2023. PMID: 36458547 Free PMC article. Review.
Cited by
-
Editorial: Identification of phenotypically important genomic variants.Front Bioinform. 2023 Nov 10;3:1328945. doi: 10.3389/fbinf.2023.1328945. eCollection 2023. Front Bioinform. 2023. PMID: 38025396 Free PMC article. No abstract available.
-
CovidTGI: A tool to investigate the temporal genetic instability of SARS-CoV-2 variants.iScience. 2025 Mar 28;28(4):112315. doi: 10.1016/j.isci.2025.112315. eCollection 2025 Apr 18. iScience. 2025. PMID: 40264800 Free PMC article.
-
Systematic analysis of SARS-CoV-2 Omicron subvariants' impact on B and T cell epitopes.PLoS One. 2024 Sep 19;19(9):e0307873. doi: 10.1371/journal.pone.0307873. eCollection 2024. PLoS One. 2024. PMID: 39298436 Free PMC article.
References
-
- World Health Organization . Tracking SARS-CoV-2 Variants. https://www.who.int/activities/tracking-SARS-CoV-2-variants (28 February 2023, date last accessed).
-
- CoV-lineages. Pango designation. https://github.com/cov-lineages/pango-designation (28 February 2023, date last accessed).
-
- Virological.org forum . https://virological.org/ (28 February 2023, date last accessed).

