Cardiolipin drives the catalytic activity of GPX4 on membranes: Insights from the R152H mutant
- PMID: 37413766
- PMCID: PMC10345155
- DOI: 10.1016/j.redox.2023.102806
Cardiolipin drives the catalytic activity of GPX4 on membranes: Insights from the R152H mutant
Abstract
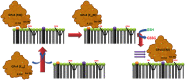
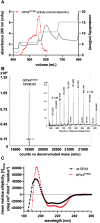
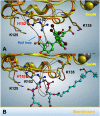
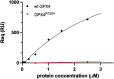
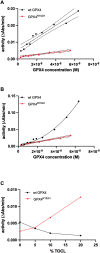
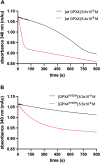
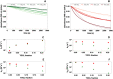
The aim of this study was to examine, in biochemical detail, the functional role of the Arg152 residue in the selenoprotein Glutathione Peroxidase 4 (GPX4), whose mutation to His is involved in Sedaghatian-type Spondylometaphyseal Dysplasia (SSMD). Wild-type and mutated recombinant enzymes with selenopcysteine (Sec) at the active site, were purified and structurally characterized to investigate the impact of the R152H mutation on enzymatic function. The mutation did not affect the peroxidase reaction's catalytic mechanism, and the kinetic parameters were qualitatively similar between the wild-type enzyme and the mutant when mixed micelles and monolamellar liposomes containing phosphatidylcholine and its hydroperoxide derivatives were used as substrate. However, in monolamellar liposomes also containing cardiolipin, which binds to a cationic area near the active site of GPX4, including residue R152, the wild-type enzyme showed a non-canonical dependency of the reaction rate on the concentration of both enzyme and membrane cardiolipin. To explain this oddity, a minimal model was developed encompassing the kinetics of both the enzyme interaction with the membrane and the catalytic peroxidase reaction. Computational fitting of experimental activity recordings showed that the wild-type enzyme was surface-sensing and prone to "positive feedback" in the presence of cardiolipin, indicating a positive cooperativity. This feature was minimal, if any, in the mutant. These findings suggest that GPX4 physiology in cardiolipin containing mitochondria is unique, and emerges as a likely target of the pathological dysfunction in SSMD.
Keywords: Cardiolipins; GPX4; Sedaghatian-type spondylometaphyseal dysplasia; Selenium.
Copyright © 2023 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no competing financial interests or personal relationships that could have influenced the work reported in this paper.
Figures

References
-
- Ursini F., Maiorino M., Valente M., Ferri L., Gregolin C. Purification from pig liver of a protein which protects liposomes and biomembranes from peroxidative degradation and exhibits glutathione peroxidase activity on phosphatidylcholine hydroperoxides. Biochim. Biophys. Acta. 1982;710(2):197–211. doi: 10.1016/0005-2760(82)90150-3. - DOI - PubMed
-
- Daolio S., Traldi P., Ursini F., Maiorino M., Gregolin C. Evidence of peroxidase activity of the peroxidation inhibiting protein on dilinoleyl phosphatidylcholine hydroperoxide as obtained in direct electron impact conditions. Biomed. Mass Spectrom. 1983;10(9):499–504.
-
- Yang W.S., SriRamaratnam R., Welsch M.E., Shimada K., Skouta R., Viswanathan V.S., Cheah J.H., Clemons P.A., Shamji A.F., Clish C.B., Brown L.M., Girotti A.W., Cornish V.W., Schreiber S.L., Stockwell B.R. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156(1–2):317–331. doi: 10.1016/j.cell.2013.12.010. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

