Characterizing control of memory CD8 T cell differentiation by BTB-ZF transcription factor Zbtb20
- PMID: 37414528
- PMCID: PMC10326419
- DOI: 10.26508/lsa.202201683
Characterizing control of memory CD8 T cell differentiation by BTB-ZF transcription factor Zbtb20
Abstract
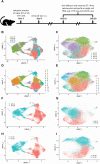
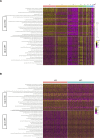
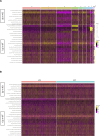
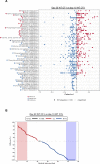
Members of the BTB-ZF transcription factor family regulate the immune system. Our laboratory identified that family member Zbtb20 contributes to the differentiation, recall responses, and metabolism of CD8 T cells. Here, we report a characterization of the transcriptional and epigenetic signatures controlled by Zbtb20 at single-cell resolution during the effector and memory phases of the CD8 T cell response. Without Zbtb20, transcriptional programs associated with memory CD8 T cell formation were up-regulated throughout the CD8 T response. A signature of open chromatin was associated with genes controlling T cell activation, consistent with the known impact on differentiation. In addition, memory CD8 T cells lacking Zbtb20 were characterized by open chromatin regions with overrepresentation of AP-1 transcription factor motifs and elevated RNA- and protein-level expressions of the corresponding AP-1 components. Finally, we describe motifs and genomic annotations from the DNA targets of Zbtb20 in CD8 T cells identified by cleavage under targets and release under nuclease (CUT&RUN). Together, these data establish the transcriptional and epigenetic networks contributing to the control of CD8 T cell responses by Zbtb20.
© 2023 Preiss et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures











Similar articles
-
Zbtb20 Restrains CD8 T Cell Immunometabolism and Restricts Memory Differentiation and Antitumor Immunity.J Immunol. 2020 Nov 15;205(10):2649-2666. doi: 10.4049/jimmunol.2000459. Epub 2020 Sep 30. J Immunol. 2020. PMID: 32998985 Free PMC article.
-
The BTB-ZF transcription factor Zbtb20 is driven by Irf4 to promote plasma cell differentiation and longevity.J Exp Med. 2014 May 5;211(5):827-40. doi: 10.1084/jem.20131831. Epub 2014 Apr 7. J Exp Med. 2014. PMID: 24711583 Free PMC article.
-
E2A-regulated epigenetic landscape promotes memory CD8 T cell differentiation.Proc Natl Acad Sci U S A. 2021 Apr 20;118(16):e2013452118. doi: 10.1073/pnas.2013452118. Proc Natl Acad Sci U S A. 2021. PMID: 33859041 Free PMC article.
-
Regulation of effector and memory CD8 + T cell differentiation: a focus on orphan nuclear receptor NR4A family, transcription factor, and metabolism.Immunol Res. 2023 Jun;71(3):314-327. doi: 10.1007/s12026-022-09353-1. Epub 2022 Dec 26. Immunol Res. 2023. PMID: 36571657 Review.
-
Epigenetics and CD8+ T cell memory.Immunol Rev. 2022 Jan;305(1):77-89. doi: 10.1111/imr.13057. Epub 2021 Dec 18. Immunol Rev. 2022. PMID: 34923638 Review.
Cited by
-
β-Catenin in Dendritic Cells Negatively Regulates CD8 T Cell Immune Responses through the Immune Checkpoint Molecule Tim-3.Vaccines (Basel). 2024 Apr 25;12(5):460. doi: 10.3390/vaccines12050460. Vaccines (Basel). 2024. PMID: 38793711 Free PMC article.
-
Zinc Finger and BTB Domain-Containing 20: A Newly Emerging Player in Pathogenesis and Development of Human Cancers.Biomolecules. 2024 Feb 4;14(2):192. doi: 10.3390/biom14020192. Biomolecules. 2024. PMID: 38397429 Free PMC article. Review.
-
GSK-3β in Dendritic Cells Exerts Opposite Functions in Regulating Cross-Priming and Memory CD8 T Cell Responses Independent of β-Catenin.Vaccines (Basel). 2024 Sep 10;12(9):1037. doi: 10.3390/vaccines12091037. Vaccines (Basel). 2024. PMID: 39340067 Free PMC article.
References
-
- Andrews S (2010) FastQC: A Quality Control Tool for High Throughput Sequence Data. Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
