Important role of endogenous microbial symbionts of fish gills in the challenging but highly biodiverse Amazonian blackwaters
- PMID: 37414754
- PMCID: PMC10326040
- DOI: 10.1038/s41467-023-39461-x
Important role of endogenous microbial symbionts of fish gills in the challenging but highly biodiverse Amazonian blackwaters
Abstract
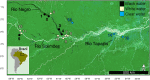
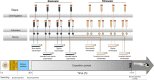
Amazonian blackwaters are extremely biodiverse systems containing some of Earth's most naturally acidic, dissolved organic carbon -rich and ion-poor waters. Physiological adaptations of fish facing these ionoregulatory challenges are unresolved but could involve microbially-mediated processes. Here, we characterize the physiological response of 964 fish-microbe systems from four blackwater Teleost species along a natural hydrochemical gradient, using dual RNA-Seq and 16 S rRNA of gill samples. We find that host transcriptional responses to blackwaters are species-specific, but occasionally include the overexpression of Toll-receptors and integrins associated to interkingdom communication. Blackwater gill microbiomes are characterized by a transcriptionally-active betaproteobacterial cluster potentially interfering with epithelial permeability. We explore further blackwater fish-microbe interactions by analyzing transcriptomes of axenic zebrafish larvae exposed to sterile, non-sterile and inverted (non-native bacterioplankton) blackwater. We find that axenic zebrafish survive poorly when exposed to sterile/inverted blackwater. Overall, our results suggest a critical role for endogenous symbionts in blackwater fish physiology.
© 2023. Crown.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Holland A, et al. Effect of key water quality variables on macroinvertebrate and fish communities within naturally acidic wallum streams. Mar. Freshw. Res. 2014;66:50. doi: 10.1071/MF13285. - DOI
-
- Sabater F, Meyer J, Edwards R. Longitudinal patterns of dissolved organic carbon concentration and suspended bacterial density along a blackwater river. Biogeochemistry. 1993;21:73–93. doi: 10.1007/BF00000872. - DOI
-
- Val, A. L., & Almeida‐Val, V. M. F. Fishes of the Amazon and their environment: physiological and biochemical features (Springer, Berlin, 1995).
-
- Walker, I., & Henderson, P. A. Ecophysiological aspects of Amazonian blackwater litterbank fish communities. In: Val, A. L., Almeida‐Val, V. M. F., & Randall, D. J. (Eds.). Physiology and Biochemistry of Fishes of the Amazon (Instituto Nacional de Pesquisas da Amazonia, Manaus, 1996).
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

