Genome-wide mapping of cancer dependency genes and genetic modifiers of chemotherapy in high-risk hepatoblastoma
- PMID: 37414763
- PMCID: PMC10326052
- DOI: 10.1038/s41467-023-39717-6
Genome-wide mapping of cancer dependency genes and genetic modifiers of chemotherapy in high-risk hepatoblastoma
Abstract
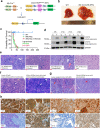
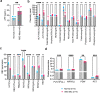
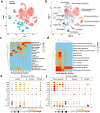
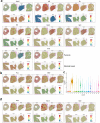
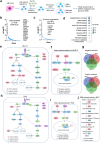
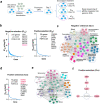
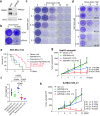
A lack of relevant genetic models and cell lines hampers our understanding of hepatoblastoma pathogenesis and the development of new therapies for this neoplasm. Here, we report an improved MYC-driven hepatoblastoma-like murine model that recapitulates the pathological features of embryonal type of hepatoblastoma, with transcriptomics resembling the high-risk gene signatures of the human disease. Single-cell RNA-sequencing and spatial transcriptomics identify distinct subpopulations of hepatoblastoma cells. After deriving cell lines from the mouse model, we map cancer dependency genes using CRISPR-Cas9 screening and identify druggable targets shared with human hepatoblastoma (e.g., CDK7, CDK9, PRMT1, PRMT5). Our screen also reveals oncogenes and tumor suppressor genes in hepatoblastoma that engage multiple, druggable cancer signaling pathways. Chemotherapy is critical for human hepatoblastoma treatment. A genetic mapping of doxorubicin response by CRISPR-Cas9 screening identifies modifiers whose loss-of-function synergizes with (e.g., PRKDC) or antagonizes (e.g., apoptosis genes) the effect of chemotherapy. The combination of PRKDC inhibition and doxorubicin-based chemotherapy greatly enhances therapeutic efficacy. These studies provide a set of resources including disease models suitable for identifying and validating potential therapeutic targets in human high-risk hepatoblastoma.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Lau CS, Mahendraraj K, Chamberlain RS. Hepatocellular carcinoma in the pediatric population: a population based clinical outcomes study involving 257 patients from the surveillance, epidemiology, and end result (SEER) database (1973–2011) HPB Surg. 2015;2015:670728. doi: 10.1155/2015/670728. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

