Dynamic de novo heterochromatin assembly and disassembly at replication forks ensures fork stability
- PMID: 37414849
- PMCID: PMC10344782
- DOI: 10.1038/s41556-023-01167-z
Dynamic de novo heterochromatin assembly and disassembly at replication forks ensures fork stability
Abstract
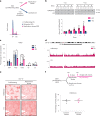
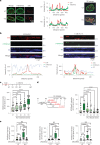
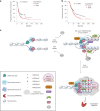
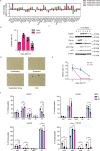
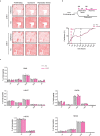
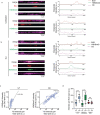
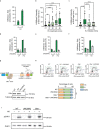
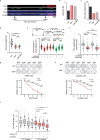
Chromatin is dynamically reorganized when DNA replication forks are challenged. However, the process of epigenetic reorganization and its implication for fork stability is poorly understood. Here we discover a checkpoint-regulated cascade of chromatin signalling that activates the histone methyltransferase EHMT2/G9a to catalyse heterochromatin assembly at stressed replication forks. Using biochemical and single molecule chromatin fibre approaches, we show that G9a together with SUV39h1 induces chromatin compaction by accumulating the repressive modifications, H3K9me1/me2/me3, in the vicinity of stressed replication forks. This closed conformation is also favoured by the G9a-dependent exclusion of the H3K9-demethylase JMJD1A/KDM3A, which facilitates heterochromatin disassembly upon fork restart. Untimely heterochromatin disassembly from stressed forks by KDM3A enables PRIMPOL access, triggering single-stranded DNA gap formation and sensitizing cells towards chemotherapeutic drugs. These findings may help in explaining chemotherapy resistance and poor prognosis observed in patients with cancer displaying elevated levels of G9a/H3K9me3.
© 2023. The Author(s).
Conflict of interest statement
A.G. is co-founder and CSO in Ankrin Therapeutics. N.T., V.G. and R.K. report an international patent filed under PCT/NL2023/050120 for ChromStretch technology. No other authors have competing interests.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

