Class 3 PI3K coactivates the circadian clock to promote rhythmic de novo purine synthesis
- PMID: 37414850
- PMCID: PMC10344785
- DOI: 10.1038/s41556-023-01171-3
Class 3 PI3K coactivates the circadian clock to promote rhythmic de novo purine synthesis
Abstract
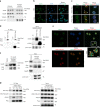
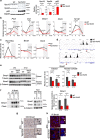
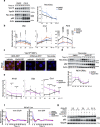
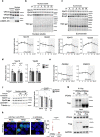
Metabolic demands fluctuate rhythmically and rely on coordination between the circadian clock and nutrient-sensing signalling pathways, yet mechanisms of their interaction remain not fully understood. Surprisingly, we find that class 3 phosphatidylinositol-3-kinase (PI3K), known best for its essential role as a lipid kinase in endocytosis and lysosomal degradation by autophagy, has an overlooked nuclear function in gene transcription as a coactivator of the heterodimeric transcription factor and circadian driver Bmal1-Clock. Canonical pro-catabolic functions of class 3 PI3K in trafficking rely on the indispensable complex between the lipid kinase Vps34 and regulatory subunit Vps15. We demonstrate that although both subunits of class 3 PI3K interact with RNA polymerase II and co-localize with active transcription sites, exclusive loss of Vps15 in cells blunts the transcriptional activity of Bmal1-Clock. Thus, we establish non-redundancy between nuclear Vps34 and Vps15, reflected by the persistent nuclear pool of Vps15 in Vps34-depleted cells and the ability of Vps15 to coactivate Bmal1-Clock independently of its complex with Vps34. In physiology we find that Vps15 is required for metabolic rhythmicity in liver and, unexpectedly, it promotes pro-anabolic de novo purine nucleotide synthesis. We show that Vps15 activates the transcription of Ppat, a key enzyme for the production of inosine monophosphate, a central metabolic intermediate for purine synthesis. Finally, we demonstrate that in fasting, which represses clock transcriptional activity, Vps15 levels are decreased on the promoters of Bmal1 targets, Nr1d1 and Ppat. Our findings open avenues for establishing the complexity for nuclear class 3 PI3K signalling for temporal regulation of energy homeostasis.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures







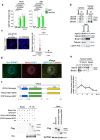



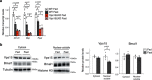
References
Publication types
MeSH terms
Substances
Grants and funding
- ERC-CoG-MetaboSENS-819543/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 European Research Council (H2020 Excellent Science - European Research Council)
- ANR-JCJC-NUTRISENSPIK-16-CE14-0029; MuscLY/Agence Nationale de la Recherche (French National Research Agency)
- ANR-21-CE14-0056-02-MuscLY/Agence Nationale de la Recherche (French National Research Agency)
- ANR_21_PRRD-0002-01/Agence Nationale de la Recherche (French National Research Agency)
- MuscLY/Agence Nationale de la Recherche (French National Research Agency)
- PHC PROCOPE/Campus France (Agence Française pour la Promotion de l'Enseignement Supérieur, l'Accueil et la Mobilité Internationale)
- PHC GERMAINE DE STAEL/Campus France (Agence Française pour la Promotion de l'Enseignement Supérieur, l'Accueil et la Mobilité Internationale)
- #23101/AFM-Téléthon (French Muscular Dystrophy Association)
- 310030_184708/1/Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung (Swiss National Science Foundation)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

