A prognostic risk score for development and spread of chronic pain
- PMID: 37414898
- PMCID: PMC10353938
- DOI: 10.1038/s41591-023-02430-4
A prognostic risk score for development and spread of chronic pain
Abstract
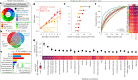
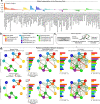
Chronic pain is a complex condition influenced by a combination of biological, psychological and social factors. Using data from the UK Biobank (n = 493,211), we showed that pain spreads from proximal to distal sites and developed a biopsychosocial model that predicted the number of coexisting pain sites. This data-driven model was used to identify a risk score that classified various chronic pain conditions (area under the curve (AUC) 0.70-0.88) and pain-related medical conditions (AUC 0.67-0.86). In longitudinal analyses, the risk score predicted the development of widespread chronic pain, the spreading of chronic pain across body sites and high-impact pain about 9 years later (AUC 0.68-0.78). Key risk factors included sleeplessness, feeling 'fed-up', tiredness, stressful life events and a body mass index >30. A simplified version of this score, named the risk of pain spreading, obtained similar predictive performance based on six simple questions with binarized answers. The risk of pain spreading was then validated in the Northern Finland Birth Cohort (n = 5,525) and the PREVENT-AD cohort (n = 178), obtaining comparable predictive performance. Our findings show that chronic pain conditions can be predicted from a common set of biopsychosocial factors, which can aid in tailoring research protocols, optimizing patient randomization in clinical trials and improving pain management.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures














Comment in
-
A risk score for pain outcomes.Nat Neurosci. 2023 Aug;26(8):1319. doi: 10.1038/s41593-023-01411-7. Nat Neurosci. 2023. PMID: 37537349 No abstract available.
References
-
- Todd KH, et al. Pain in the emergency department: results of the pain and emergency medicine initiative (PEMI) multicenter study. J. Pain. 2007;8:460–466. - PubMed
-
- Chou R, Shekelle P. Will this patient develop persistent disabling low back pain? JAMA. 2010;303:1295–1302. - PubMed
-
- Gatchel RJ, Peng YB, Peters ML, Fuchs PN, Turk DC. The biopsychosocial approach to chronic pain: scientific advances and future directions. Psychol. Bull. 2007;133:581. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

