A multi-ancestry polygenic risk score improves risk prediction for coronary artery disease
- PMID: 37414900
- PMCID: PMC10353935
- DOI: 10.1038/s41591-023-02429-x
A multi-ancestry polygenic risk score improves risk prediction for coronary artery disease
Abstract
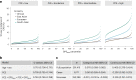
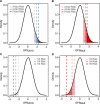
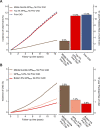
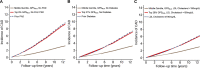
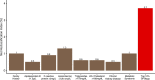
Identification of individuals at highest risk of coronary artery disease (CAD)-ideally before onset-remains an important public health need. Prior studies have developed genome-wide polygenic scores to enable risk stratification, reflecting the substantial inherited component to CAD risk. Here we develop a new and significantly improved polygenic score for CAD, termed GPSMult, that incorporates genome-wide association data across five ancestries for CAD (>269,000 cases and >1,178,000 controls) and ten CAD risk factors. GPSMult strongly associated with prevalent CAD (odds ratio per standard deviation 2.14, 95% confidence interval 2.10-2.19, P < 0.001) in UK Biobank participants of European ancestry, identifying 20.0% of the population with 3-fold increased risk and conversely 13.9% with 3-fold decreased risk as compared with those in the middle quintile. GPSMult was also associated with incident CAD events (hazard ratio per standard deviation 1.73, 95% confidence interval 1.70-1.76, P < 0.001), identifying 3% of healthy individuals with risk of future CAD events equivalent to those with existing disease and significantly improving risk discrimination and reclassification. Across multiethnic, external validation datasets inclusive of 33,096, 124,467, 16,433 and 16,874 participants of African, European, Hispanic and South Asian ancestry, respectively, GPSMult demonstrated increased strength of associations across all ancestries and outperformed all available previously published CAD polygenic scores. These data contribute a new GPSMult for CAD to the field and provide a generalizable framework for how large-scale integration of genetic association data for CAD and related traits from diverse populations can meaningfully improve polygenic risk prediction.
© 2023. The Author(s).
Conflict of interest statement
S.A. has served as a scientific advisor to Third Rock Ventures. A.C.F. is a co-founder of Goodpath and reports a grant from Abbott Vascular. P.T.E. receives sponsored research support from Bayer AG and IBM Research; he has also served on advisory boards or consulted for Bayer AG, MyoKardia and Novartis. A.S.B. reports institutional grants from AstraZeneca, Bayer, Biogen, BioMarin, Bioverativ, Novartis, Regeneron and Sanofi. P.N. reports research grants from Allelica, Apple, Amgen, Boston Scientific, Genentech/Roche and Novartis, personal fees from Allelica, Apple, AstraZeneca, Blackstone Life Sciences, Foresite Labs, Genentech/Roche, GV, HeartFlow, Magnet Biomedicine and Novartis, scientific advisory board membership of Esperion Therapeutics, Preciseli and TenSixteen Bio, scientific co-founder of TenSixteen Bio, equity in Preciseli and TenSixteen Bio, and spousal employment at Vertex Pharmaceuticals, all unrelated to the present work. A.V.K. is an employee of Verve Therapeutics; has served as a scientific advisor to Amgen, Novartis, Silence Therapeutics, Korro Bio, Veritas International, Color Health, Third Rock Ventures, Illumina, Ambry and Foresite Labs; holds equity in Verve Therapeutics, Color Health and Foresite Labs; and is listed as a co-inventor on patent applications related to assessment and mitigation of risk associated with perturbations in body fat distribution. The remaining authors declare no competing interests.
Figures
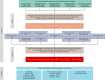
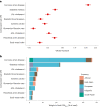
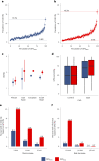

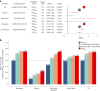

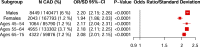

References
-
- Goff David C, et al. 2013 ACC/AHA guideline on the assessment of cardiovascular risk. Circulation. 2014;129:S49–S73. - PubMed
Publication types
MeSH terms
Grants and funding
- 210561/Z/18/Z/WT_/Wellcome Trust/United Kingdom
- RG/18/13/33946/BHF_/British Heart Foundation/United Kingdom
- I01 BX004821/BX/BLRD VA/United States
- WT102627/WT_/Wellcome Trust/United Kingdom
- R01 HL092577/HL/NHLBI NIH HHS/United States
- CH/12/2/29428/BHF_/British Heart Foundation/United Kingdom
- R01 HL164629/HL/NHLBI NIH HHS/United States
- I01 BX003362/BX/BLRD VA/United States
- M009017/MRC_/Medical Research Council/United Kingdom
- K08 HL161448/HL/NHLBI NIH HHS/United States
- R01 HL148565/HL/NHLBI NIH HHS/United States
- U01 HG011719/HG/NHGRI NIH HHS/United States
- MR/X009920/1/MRC_/Medical Research Council/United Kingdom
- WT210561/WT_/Wellcome Trust/United Kingdom
- R01 HL157635/HL/NHLBI NIH HHS/United States
- K08 HL153937/HL/NHLBI NIH HHS/United States
- MR/X009777/1/MRC_/Medical Research Council/United Kingdom
- R01 HL148050/HL/NHLBI NIH HHS/United States
- K08 HG010155/HG/NHGRI NIH HHS/United States
- KL2 TR003143/TR/NCATS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

