The ribosomal protein S6 kinase alpha-1 (RPS6KA1) induces resistance to venetoclax/azacitidine in acute myeloid leukemia
- PMID: 37414921
- PMCID: PMC10400424
- DOI: 10.1038/s41375-023-01951-8
The ribosomal protein S6 kinase alpha-1 (RPS6KA1) induces resistance to venetoclax/azacitidine in acute myeloid leukemia
Abstract
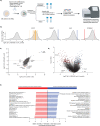
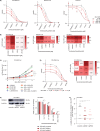
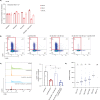
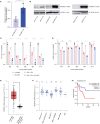
Venetoclax/azacitidine combination therapy is effective in acute myeloid leukemia (AML) and tolerable for older, multimorbid patients. Despite promising response rates, many patients do not achieve sustained remission or are upfront refractory. Identification of resistance mechanisms and additional therapeutic targets represent unmet clinical needs. By using a genome-wide CRISPR/Cas9 library screen targeting 18,053 protein- coding genes in a human AML cell line, various genes conferring resistance to combined venetoclax/azacitidine treatment were identified. The ribosomal protein S6 kinase A1 (RPS6KA1) was among the most significantly depleted sgRNA-genes in venetoclax/azacitidine- treated AML cells. Addition of the RPS6KA1 inhibitor BI-D1870 to venetoclax/azacitidine decreased proliferation and colony forming potential compared to venetoclax/azacitidine alone. Furthermore, BI-D1870 was able to completely restore the sensitivity of OCI-AML2 cells with acquired resistance to venetoclax/azacitidine. Analysis of cell surface markers revealed that RPS6KA1 inhibition efficiently targeted monocytic blast subclones as a potential source of relapse upon venetoclax/azacitidine treatment. Taken together, our results suggest RPS6KA1 as mediator of resistance towards venetoclax/azacitidine and additional RPS6KA1 inhibition as strategy to prevent or overcome resistance.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

