Human resident liver myeloid cells protect against metabolic stress in obesity
- PMID: 37414931
- PMCID: PMC10365994
- DOI: 10.1038/s42255-023-00834-7
Human resident liver myeloid cells protect against metabolic stress in obesity
Abstract
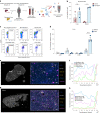
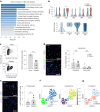
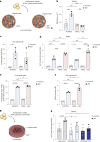
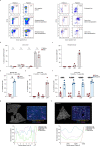
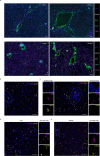
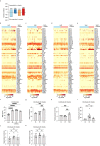
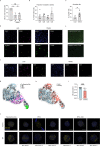
Although multiple populations of macrophages have been described in the human liver, their function and turnover in patients with obesity at high risk of developing non-alcoholic fatty liver disease (NAFLD) and cirrhosis are currently unknown. Herein, we identify a specific human population of resident liver myeloid cells that protects against the metabolic impairment associated with obesity. By studying the turnover of liver myeloid cells in individuals undergoing liver transplantation, we find that liver myeloid cell turnover differs between humans and mice. Using single-cell techniques and flow cytometry, we determine that the proportion of the protective resident liver myeloid cells, denoted liver myeloid cells 2 (LM2), decreases during obesity. Functional validation approaches using human 2D and 3D cultures reveal that the presence of LM2 ameliorates the oxidative stress associated with obese conditions. Our study indicates that resident myeloid cells could be a therapeutic target to decrease the oxidative stress associated with NAFLD.
© 2023. The Author(s).
Conflict of interest statement
V.M.L. is founder, CEO and shareholder of HepaPredict AB. In addition, V.M.L. discloses consultancy work for EnginZyme AB. The remaining authors declare no competing interests.
Figures








Comment in
-
Antioxidant liver myeloid cell population identified.Nat Rev Endocrinol. 2023 Oct;19(10):556. doi: 10.1038/s41574-023-00880-x. Nat Rev Endocrinol. 2023. PMID: 37491639 No abstract available.
References
-
- Younossi ZM. Patient-reported outcomes for patients with chronic liver disease. Clin. Gastroenterol. Hepatol. 2018;16:793–799. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

