Comparative genomics reveals insights into anuran genome size evolution
- PMID: 37415107
- PMCID: PMC10324214
- DOI: 10.1186/s12864-023-09499-8
Comparative genomics reveals insights into anuran genome size evolution
Abstract
Background: Amphibians, particularly anurans, display an enormous variation in genome size. Due to the unavailability of whole genome datasets in the past, the genomic elements and evolutionary causes of anuran genome size variation are poorly understood. To address this, we analyzed whole-genome sequences of 14 anuran species ranging in size from 1.1 to 6.8 Gb. By annotating multiple genomic elements, we investigated the genomic correlates of anuran genome size variation and further examined whether the genome size relates to habitat types.
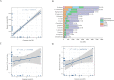
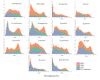
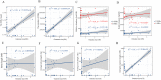
Results: Our results showed that intron expansions or contraction and Transposable Elements (TEs) diversity do not contribute significantly to genome size variation. However, the recent accumulation of transposable elements (TEs) and the lack of deletion of ancient TEs primarily accounted for the evolution of anuran genome sizes. Our study showed that the abundance and density of simple repeat sequences positively correlate with genome size. Ancestral state reconstruction revealed that genome size exhibits a taxon-specific pattern of evolution, with families Bufonidae and Pipidae experiencing extreme genome expansion and contraction events, respectively. Our result showed no relationship between genome size and habitat types, although large genome-sized species are predominantly found in humid habitats.
Conclusions: Overall, our study identified the genomic element and their evolutionary dynamics accounting for anuran genome size variation, thus paving a path to a greater understanding of the size evolution of the genome in amphibians.
Keywords: Anurans; Genome size variation; Simple repeat sequences; Transposable elements.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

